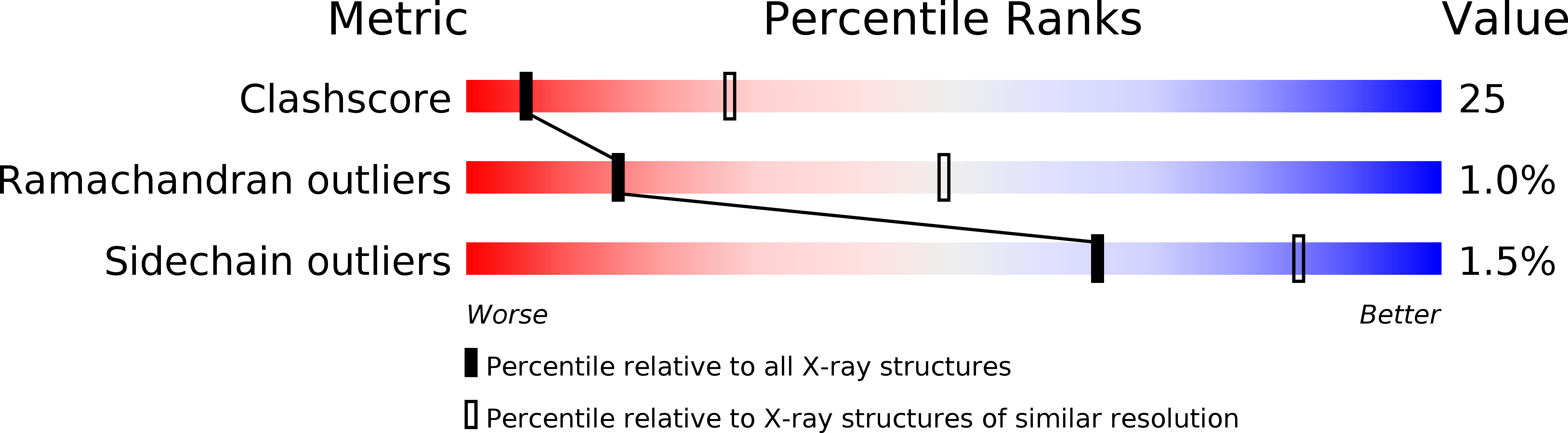Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
Howitt, J., Bewley, M.C., Graziano, V., Flanagan, J.M., Freimuth, P.(2003) J Biol Chem 278: 26208-26215
- PubMed: 12716886
- DOI: https://doi.org/10.1074/jbc.M301492200
- Primary Citation of Related Structures:
1P69, 1P6A - PubMed Abstract:
The majority of adenovirus serotypes can bind to the coxsackievirus and adenovirus receptor (CAR) on human cells despite only limited conservation of the amino acid residues that comprise the receptor-binding sites of these viruses. Using a fluorescence anisotropy-based assay, we determined that the recombinant knob domain of the fiber protein from adenovirus serotype (Ad) 2 binds the soluble, N-terminal domain (domain 1 (D1)) of CAR with 8-fold greater affinity than does the recombinant knob domain from Ad12. Homology modeling predicted that the increased affinity of Ad2 knob for CAR D1 could result from additional contacts within the binding interface contributed by two residues, Ser408 and Tyr477, which are not conserved in the Ad12 knob. Consistent with this structural model, substitution of serine and tyrosine for the corresponding residues in the Ad12 knob (P417S and S489Y) increased the binding affinity by 4- and 8-fold, respectively, whereas the double mutation increased binding affinity 10-fold. X-ray structure analysis of Ad12 knob mutants P417S and S489Y indicated that both substituted residues potentially could form additional hydrogen bonds across the knob-CAR interface. Structural changes resulting from these mutations were highly localized, implying that the high tolerance for surface variation conferred by the stable knob scaffold can minimize the impact of antigenic drift on binding specificity and affinity during evolution of virus serotypes. Our results suggest that the interaction of knob domains from different adenovirus serotypes with CAR D1 can be accurately modeled using the Ad12 knob-CAR D1 crystal structure as a template.
Organizational Affiliation:
Biology Department, Brookhaven National Laboratory, Upton, New York 11973, USA.