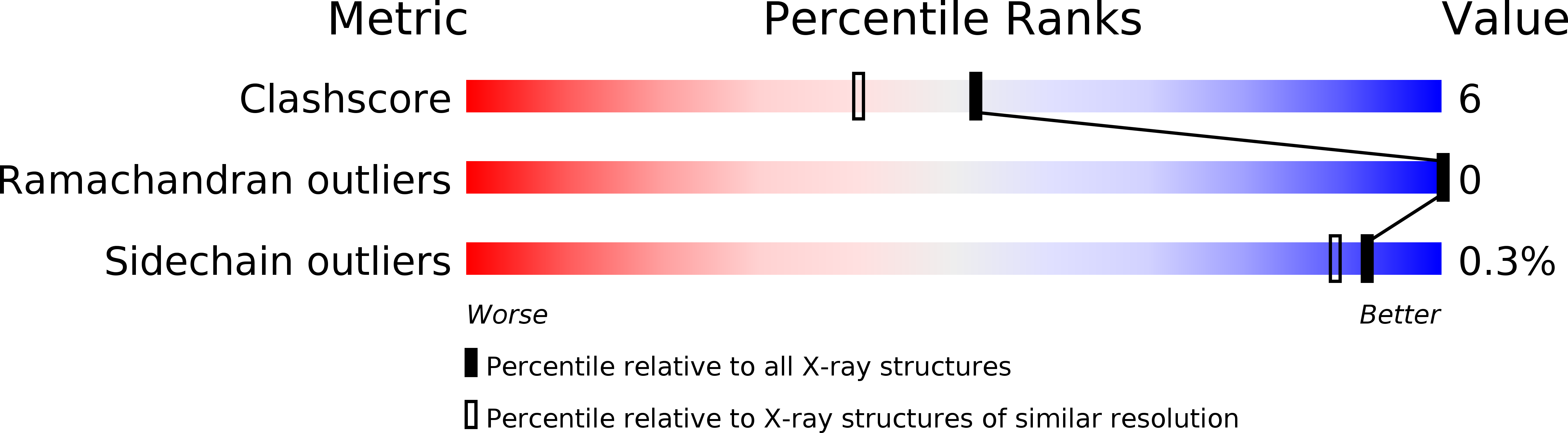Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
Golan, G., Shallom, D., Teplitsky, A., Zaide, G., Shulami, S., Baasov, T., Stojanoff, V., Thompson, A., Shoham, Y., Shoham, G.(2004) J Biol Chem 279: 3014-3024
- PubMed: 14573597
- DOI: https://doi.org/10.1074/jbc.M310098200
- Primary Citation of Related Structures:
1K9D, 1K9E, 1K9F, 1L8N, 1MQP, 1MQQ, 1MQR - PubMed Abstract:
Alpha-glucuronidases cleave the alpha-1,2-glycosidic bond between 4-O-methyl-d-glucuronic acid and short xylooligomers as part of the hemicellulose degradation system. To date, all of the alpha-glucuronidases are classified as family 67 glycosidases, which catalyze the hydrolysis via the investing mechanism. Here we describe several high resolution crystal structures of the alpha-glucuronidase (AguA) from Geobacillus stearothermophilus, in complex with its substrate and products. In the complex of AguA with the intact substrate, the 4-O-methyl-d-glucuronic acid sugar ring is distorted into a half-chair conformation, which is closer to the planar conformation required for the oxocarbenium ion-like transition state structure. In the active site, a water molecule is coordinated between two carboxylic acids, in an appropriate position to act as a nucleophile. From the structural data it is likely that two carboxylic acids, Asp(364) and Glu(392), activate together the nucleophilic water molecule. The loop carrying the catalytic general acid Glu(285) cannot be resolved in some of the structures but could be visualized in its "open" and "closed" (catalytic) conformations in other structures. The protonated state of Glu(285) is presumably stabilized by its proximity to the negative charge of the substrate, representing a new variation of substrate-assisted catalysis mechanism.
Organizational Affiliation:
Department of Inorganic Chemistry, The Hebrew University of Jerusalem, Jerusalem 91904, Israel.