Differences in anionic inhibition of human carbonic anhydrase I revealed from the structures of iodide and gold cyanide inhibitor complexes.
Kumar, V., Kannan, K.K., Sathyamurthi, P.(1994) Acta Crystallogr D Biol Crystallogr 50: 731-738
- PubMed: 15299369
- DOI: https://doi.org/10.1107/S0907444994001873
- Primary Citation of Related Structures:
1HUG, 1HUH - PubMed Abstract:
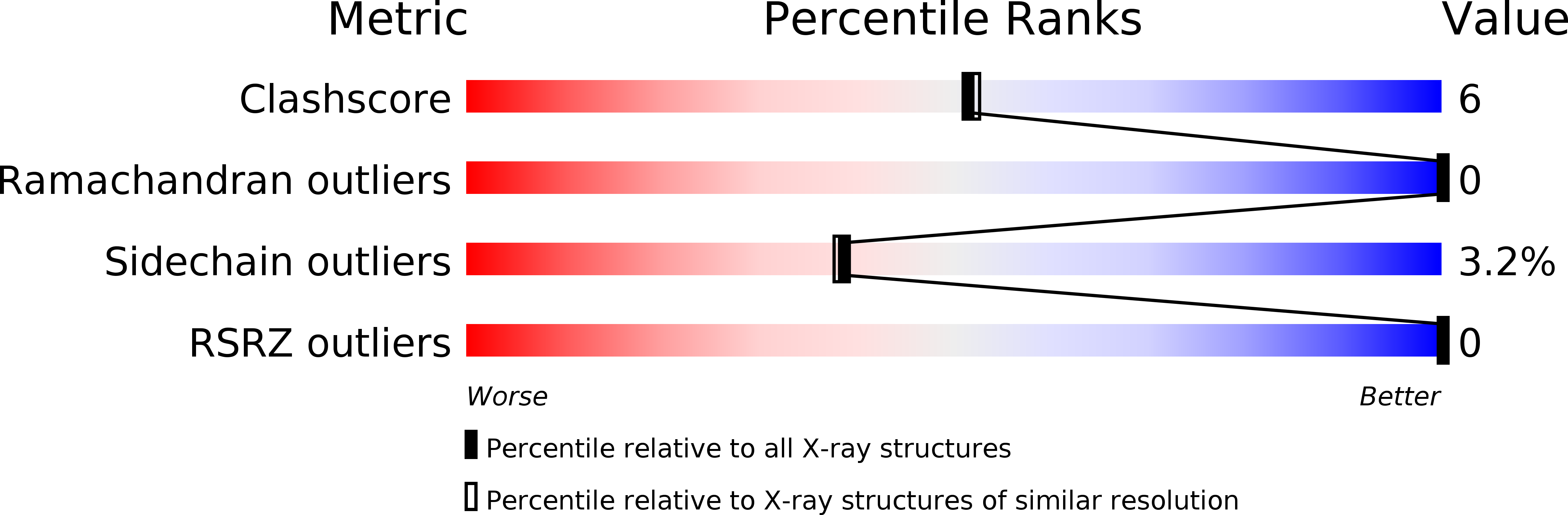
The crystal structures of two anionic inhibitor complexes of human carbonic anhydrase I (HCAI), namely, HCAI-iodide and HCAI-Au(CN)(2)(-), have been refined by the restrained least-squares method at 2.2 and 2 A nominal resolution, respectively, with good stereochemistry for the final models. The R values have improved from 30.3 to 16.6% for HCAI-iodide and from 28.8 to 17.1% for HCAI-Au(CN)(2)(-). The sites of inhibitor binding as elucidated are totally different in the two structures. The iodide anion replaces the zinc-bound H(2)O/OH(-) ligand and renders the enzyme inactive. This result confirms that the zinc-bound H(2)O/OH(-) is the activity-linked group in carbonic anhydrase enzymes. Au(CN)(2)(-) binds at a different and new site near the zinc ion, without liganding to the metal. The N atom of Au(CN)(2)(-) is within hydrogen-bonding distance of the zinc-bound H(2)O/OH(-) group which shifts by about 0.4 A away from the zinc ion in relation to its position in the native HCAI. It is proposed that the presence of the inhibitor Au(CN)(2)(-) results in a conformational reorientation of the activity-linked group, due to hydrogen-bond formation with the inhibitor, which in turn sterically hinders the binding of the substrate CO(2) molecule in the active site, leading to the inhibition of HCAI enzyme.
Organizational Affiliation:
Solid State Physics Division, Bhabha Atomic Research Centre, Bombay, India.