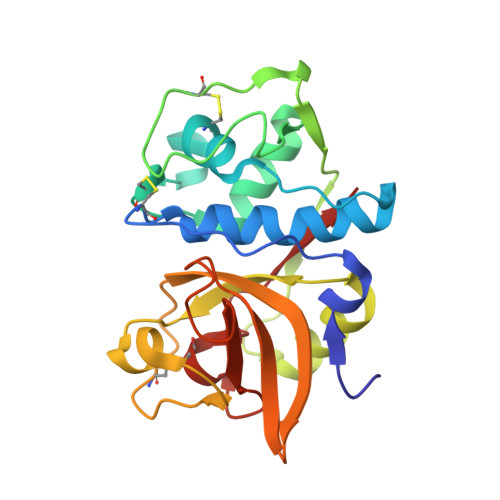
Structure of a Cys25->Ser Mutant of Human Cathepsin Cathepsin S
Turkenburg, J.P., Lamers, M.B.A.C., Brzozowski, A.M., Wright, L.M., Hubbard, R.E., Sturt, S.L., Williams, D.H.(2002) Acta Crystallogr D Biol Crystallogr 58: 451
- PubMed: 11856830
- DOI: https://doi.org/10.1107/s0907444901021825
- Primary Citation of Related Structures:
1GLO - PubMed Abstract:
Cathepsin S (EC 3.4.22.27), a cysteine proteinase of the papain superfamily, plays a critical role in the generation of a major histocompatibility complex (MHC) class II restricted T-cell response by antigen-presenting cells. Therefore, selective inhibition of this enzyme may be useful in modulating class II restricted T-cell responses in immune-related disorders such as rheumatoid arthritis, multiple sclerosis and extrinsic asthma. The three-dimensional structure at 2.2 A resolution of the active-site Cys25-->Ser mutant presented here in an unliganded state provides further insight useful for the design of selective enzyme inhibitors.
Organizational Affiliation:
York Structural Biology Laboratory, Chemistry Department, University of York, Heslington, York YO10 5DD, England.