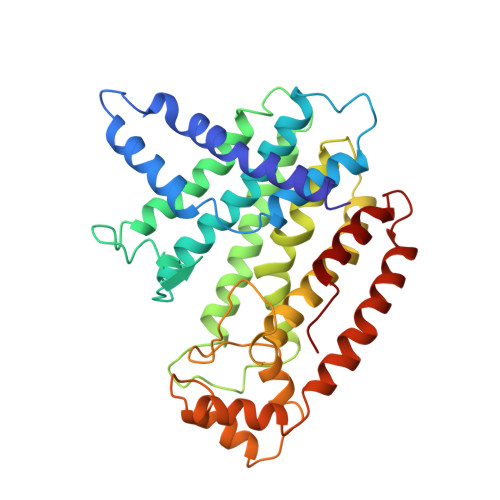
Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-A resolution.
Tarshis, L.C., Yan, M., Poulter, C.D., Sacchettini, J.C.(1994) Biochemistry 33: 10871-10877
- PubMed: 8086404
- DOI: https://doi.org/10.1021/bi00202a004
- Primary Citation of Related Structures:
1FPS - PubMed Abstract:
The synthesis of farnesyl diphosphate (FPP), a key intermediate in the isoprenoid biosynthetic pathway required for the synthesis of cholesterol and in the formation of prenylated proteins, is catalyzed by the enzyme farnesyl diphosphate synthase (FPS). The crystal structure of avian recombinant FPS, the first three-dimensional structure for any prenyltransferase, was determined to 2.6-A resolution. The enzyme exhibits a novel fold composed entirely of alpha-helices joined by connecting loops. The enzyme's most prominent structural feature is the arrangement of 10 core helices around a large central cavity. Two aspartate-rich sequences that are highly conserved among the isoprenyl diphosphate synthase family of prenyltransferases, and are essential for enzymatic activity, were found on opposite walls of this cavity, with the aspartate side chains approximately 12 A apart and facing each other. The location and metal ion binding properties of these sequences suggest that the conserved aspartate residues participate in substrate binding of catalysis.
Organizational Affiliation:
Department of Biochemistry, Albert Einstein College of Medicine, Bronx, New York 10461.