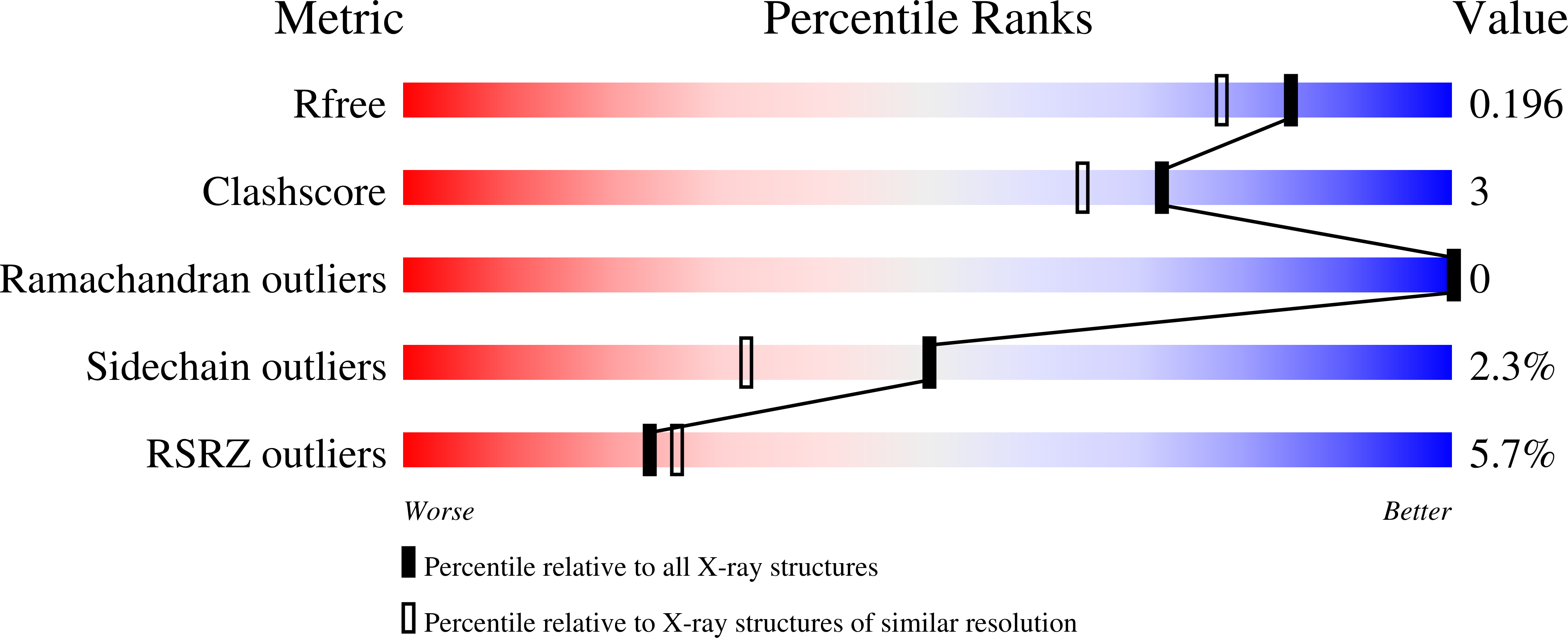
Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
van Raaij, M.J., Chouin, E., van der Zandt, H., Bergelson, J.M., Cusack, S.(2000) Structure 8: 1147-1155
- PubMed: 11080637
- DOI: https://doi.org/10.1016/s0969-2126(00)00528-1
- Primary Citation of Related Structures:
1EAJ, 1F5W - PubMed Abstract:
The coxsackievirus and adenovirus receptor (CAR) comprises two extracellular immunoglobulin domains, a transmembrane helix and a C-terminal intracellular domain. The amino-terminal immunoglobulin domain (D1) of CAR is necessary and sufficient for adenovirus binding, whereas the site of coxsackievirus attachment has not yet been localized. The normal cellular role of CAR is currently unknown, although CAR was recently proposed to function as a homophilic cell adhesion molecule. The human CAR D1 domain was bacterially expressed and crystallized. The structure was solved by molecular replacement using the structure of CAR D1 bound to the adenovirus type 12 fiber head and refined to 1.7 A resolution, including individual anisotropic temperature factors. The two CAR D1 structures are virtually identical, apart from the BC, C"D, and FG loops that are involved both in fiber head binding and homodimerization in the crystal. Analytical equilibrium ultracentrifugation shows that a dimer also exists in solution, with a dissociation constant of 16 microM. The CAR D1 domain forms homodimers in the crystal using the same GFCC'C" surface that interacts with the adenovirus fiber head. The homodimer is very similar to the CD2 D1-CD58 D1 heterodimer. CAR D1 also forms dimers in solution with a dissociation constant typical of other cell adhesion complexes. These results are consistent with reports that CAR may function physiologically as a homophilic cell adhesion molecule in the developing mouse brain. Adenovirus may thus have recruited an existing and conserved interaction surface of CAR to use for its own cell attachment.
Organizational Affiliation:
European Molecular Biology Laboratory Grenoble Outstation c/o Institut Laue Langevin BP 156 F-38042 9, Grenoble Cedex, France. m.vanraaij@chem.leidenuniv.nl