The Hairpin Structure of the (6)F1(1)F2(2)F2 Fragment from Human Fibronectin Enhances Gelatin Binding
Pickford, A.R., Smith, S.P., Staunton, D., Boyd, J., Campbell, I.D.(2001) EMBO J 20: 1519-1529
- PubMed: 11285216
- DOI: https://doi.org/10.1093/emboj/20.7.1519
- Primary Citation of Related Structures:
1E88, 1E8B - PubMed Abstract:
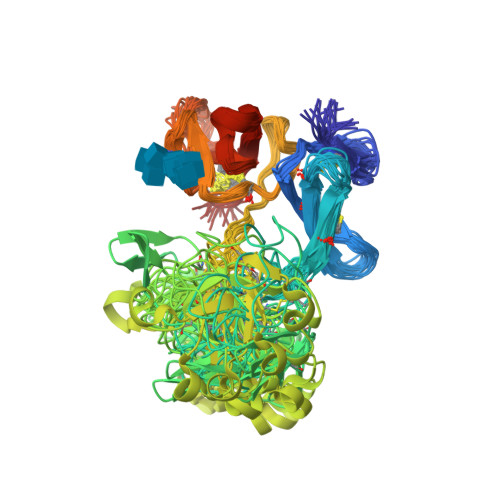
The solution structure of the (6)F1(1)F2(2)F2 fragment from the gelatin-binding region of fibronectin has been determined (Protein Data Bank entry codes 1e88 and 1e8b). The structure reveals an extensive hydrophobic interface between the non-contiguous (6)F1 and (2)F2 modules. The buried surface area between (6)F1 and (2)F2 ( approximately 870 A(2)) is the largest intermodule interface seen in fibronectin to date. The dissection of (6)F1(1)F2(2)F2 into the (6)F1(1)F2 pair and (2)F2 results in near-complete loss of gelatin-binding activity. The hairpin topology of (6)F1(1)F2(2)F2 may facilitate intramolecular contact between the matrix assembly regions flanking the gelatin-binding domain. This is the first high-resolution study to reveal a compact, globular arrangement of modules in fibronectin. This arrangement is not consistent with the view that fibronectin is simply a linear 'string of beads'.
Organizational Affiliation:
Department of Biochemistry and Oxford Centre for Molecular Sciences, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.