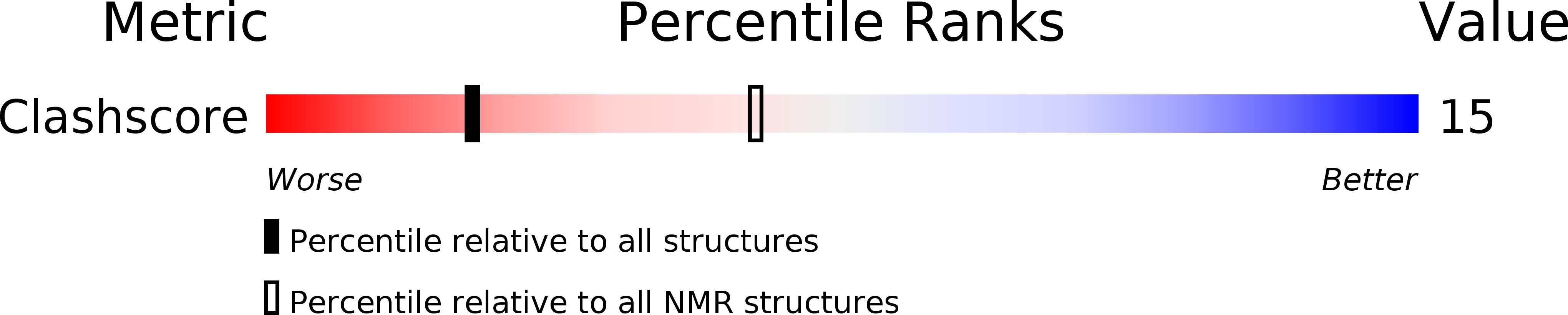
Hydration of [D(Cgc)R(Aaa)D(Tttgcg)]2
Hsu, S.-T., Chou, M.-T., Chou, S.-H., Huang, W.-C., Cheng, J.-W.(2000) J Mol Biol 295: 1129
- PubMed: 10653692
- DOI: https://doi.org/10.1006/jmbi.1999.3388
- Primary Citation of Related Structures:
1DXN - PubMed Abstract:
We have studied the hydration and dynamics of RNA C2'-OH in a DNA. RNA hybrid chimeric duplex [d(CGC)r(aaa)d(TTTGCG)](2). Long-lived water molecules with correlation time tau(c) larger than 0.3 ns were found close to the RNA adenine H2 and H1' protons in the hybrid segment. A possible long-lived water molecule was also detected close to the methyl group of 7T in the RNA-DNA junction but not to the other two thymine bases (8T and 9T). This result correlates with the structural studies that only DNA residue 7T in the RNA-DNA junction adopts an O4'-endo sugar conformation (intermediate between B-form and A-form), while the other DNA residues including 3C in the DNA-RNA junction, adopt C1'-exo or C2'-endo conformations (in the B-form domain). Based on the NOE cross-peak patterns, we have found that RNA C2'-OH tends to orient toward the O3' direction, forming a possible hydrogen bond with the 3'-phosphate group. The exchange rates for RNA C2'-OH were found to be around 5-20 s(-1), compared to 26.7(+/-13.8) s(-1) reported previously for the other DNA.RNA hybrid duplex. This slow exchange rate may be due to the narrow minor groove width of [d(CGC)r(aaa)d(TTTGCG)](2), which may trap the water molecules and restrict the dynamic motion of hydroxyl protons. The distinct hydration patterns of the RNA adenine H2 and H1' protons and the DNA 7T methyl group in the hybrid segment, as well as the orientation and dynamics of the RNA C2'-OH protons, may provide a molecular basis for further understanding the structure and recognition of DNA.RNA hybrid and chimeric duplexes.
Organizational Affiliation:
Division of Structural Biology and Biomedical Science Department of Life Science, National Tsing Hua University, Hsinchu 300, Taiwan, ROC.