Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex.
Sandler, B.H., Nikonova, L., Leal, W.S., Clardy, J.(2000) Chem Biol 7: 143-151
- PubMed: 10662696
- DOI: https://doi.org/10.1016/s1074-5521(00)00078-8
- Primary Citation of Related Structures:
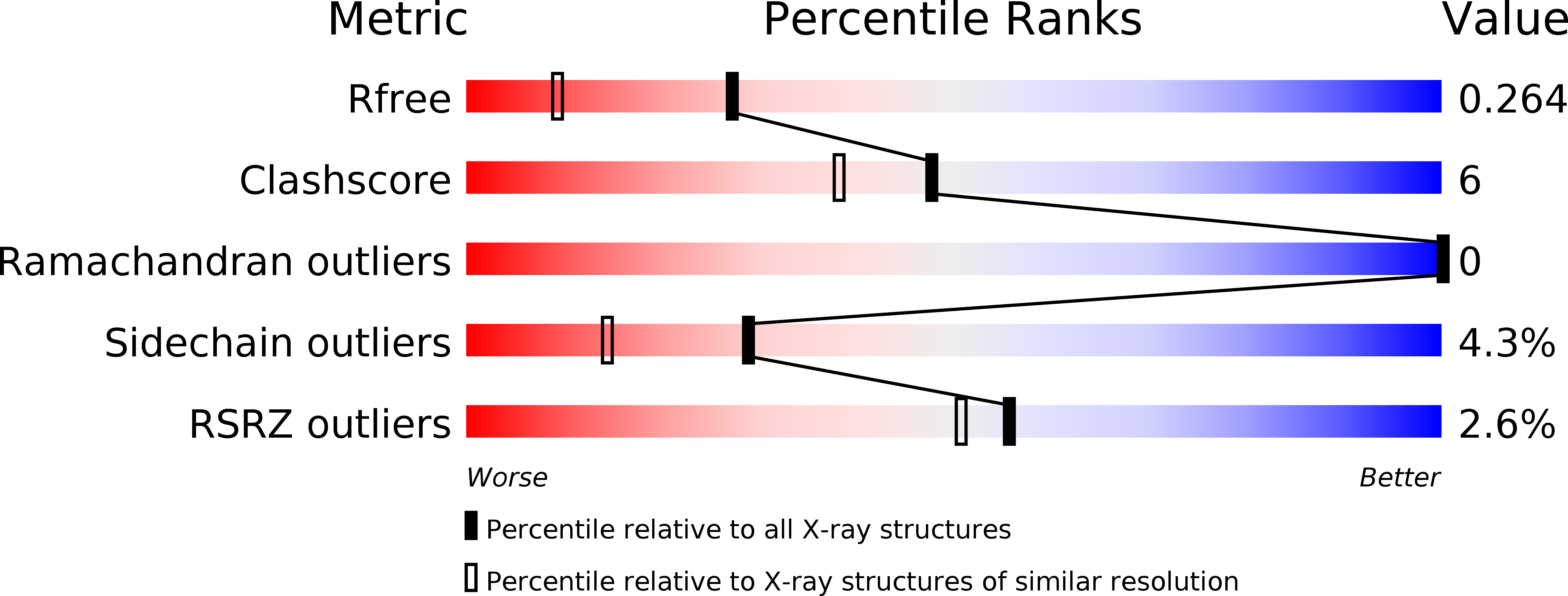
1DQE - PubMed Abstract:
Insects use volatile organic molecules to communicate messages with remarkable sensitivity and specificity. In one of the most studied systems, female silkworm moths (Bombyx mori) attract male mates with the pheromone bombykol, a volatile 16-carbon alcohol. In the male moth's antennae, a pheromone-binding protein conveys bombykol to a membrane-bound receptor on a nerve cell. The structure of the pheromone-binding protein, its binding and recognition of bombykol, and its full role in signal transduction are not known. The three-dimensional structure of the B. mori pheromone-binding protein with bound bombykol has been determined by X-ray diffraction at 1.8 A resolution. The pheromone binding protein of B. mori has six helices, and bombykol binds in a completely enclosed hydrophobic cavity formed by four antiparallel helices. Bombykol is bound in this cavity through numerous hydrophobic interactions, and sequence alignments suggest critical residues for specific pheromone binding.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853-1301, USA.