Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains.
Wilson, K.P., Shewchuk, L.M., Brennan, R.G., Otsuka, A.J., Matthews, B.W.(1992) Proc Natl Acad Sci U S A 89: 9257-9261
- PubMed: 1409631
- DOI: https://doi.org/10.1073/pnas.89.19.9257
- Primary Citation of Related Structures:
1BIA, 1BIB - PubMed Abstract:
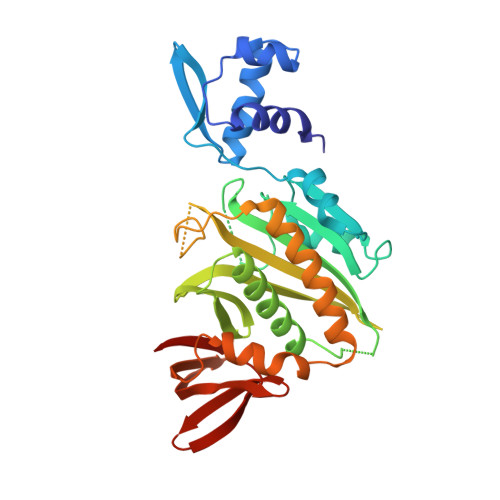
The three-dimensional structure of BirA, the repressor of the Escherichia coli biotin biosynthetic operon, has been determined by x-ray crystallography and refined to a crystallographic residual of 19.0% at 2.3-A resolution. BirA is a sequence-specific DNA-binding protein that also catalyzes the formation of biotinyl-5'-adenylate from biotin and ATP and transfers the biotin moiety to other proteins. The level of biotin biosynthetic enzymes in the cell is controlled by the amount of biotinyl-5'-adenylate, which is the BirA corepressor. The structure provides an example of a transcription factor that is also an enzyme. The structure of BirA is highly asymmetric and consists of three domains. The N-terminal domain is mostly alpha-helical, contains a helix-turn-helix DNA-binding motif, and is loosely connected to the remainder of the molecule. The central domain consists of a seven-stranded mixed beta-sheet with alpha-helices covering one face. The other side of the sheet is largely solvent-exposed and contains the active site. The C-terminal domain comprises a six-stranded, antiparallel beta-sheet sandwich. The location of biotin binding is consistent with mutations that affect enzymatic activity. A nearby loop has a sequence that has been associated with phosphate binding in other proteins. It is inferred that ATP binds in this region, adjacent to the biotin. It is proposed that the binding of corepressor to monomeric BirA may promote DNA binding by facilitating the formation of a multimeric BirA-corepressor-DNA complex. The structural details of this complex remain an open question, however.
Organizational Affiliation:
Institute of Molecular Biology, Howard Hughes Medical Institute, University of Oregon, Eugene 97403.