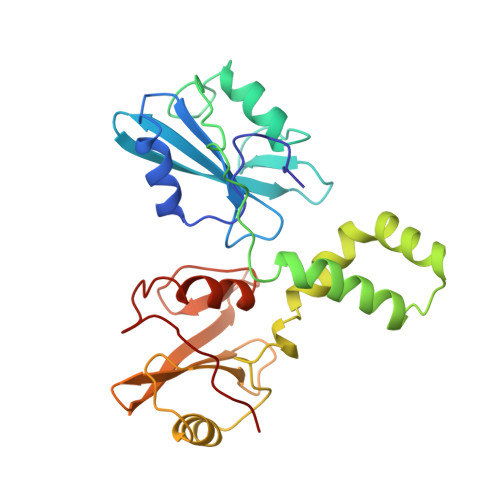
Structural basis for Syk tyrosine kinase ubiquity in signal transduction pathways revealed by the crystal structure of its regulatory SH2 domains bound to a dually phosphorylated ITAM peptide.
Futterer, K., Wong, J., Grucza, R.A., Chan, A.C., Waksman, G.(1998) J Mol Biol 281: 523-537
- PubMed: 9698567
- DOI: https://doi.org/10.1006/jmbi.1998.1964
- Primary Citation of Related Structures:
1A81 - PubMed Abstract:
The Syk family of kinases, consisting of ZAP-70 and Syk, play essential roles in a variety of immune and non-immune cells. This family of kinases is characterized by the presence of two adjacent SH2 domains which mediate their localization to the membrane through receptor encoded tyrosine phosphorylated motifs. While these two kinases share many structural and functional features, the more ubiquitous nature of Syk has suggested that this kinase may accommodate a greater variety of motifs to mediate its function. We present the crystal structure of the tandem SH2 domain of Syk complexed with a dually phosphorylated ITAM peptide. The structure was solved by multiple isomorphous replacement at 3.0 A resolution. The asymmetric unit comprises six copies of the liganded protein, revealing a surprising flexibility in the relative orientation of the two SH2 domains. The C-terminal phosphotyrosine-binding site is very different from the equivalent region of ZAP-70, suggesting that in contrast to ZAP-70, the two SH2 domains of Syk can function as independent units. The conformational flexibility and structural independence of the SH2 modules of Syk likely provides the molecular basis for the more ubiquitous involvement of Syk in a variety of signal transduction pathways.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, 660 South Euclid Av., Saint Louis, MO, 63110, USA.