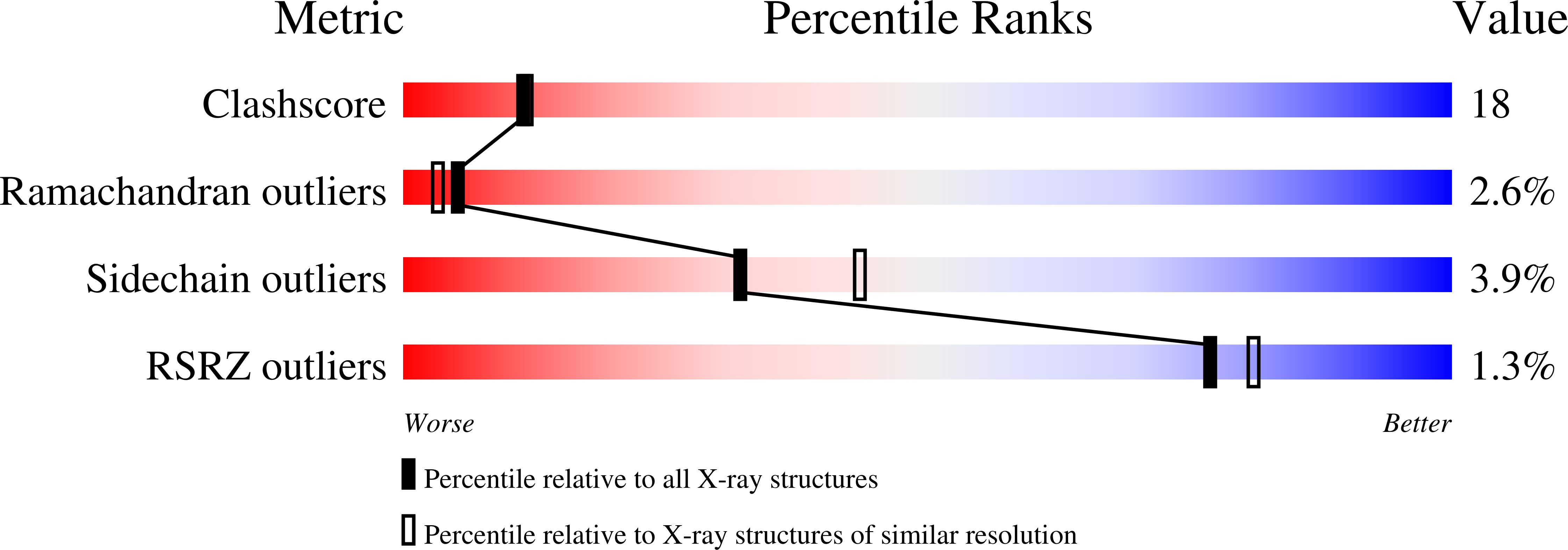
Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography.
Yu, B., Blaber, M., Gronenborn, A.M., Clore, G.M., Caspar, D.L.(1999) Proc Natl Acad Sci U S A 96: 103-108
- PubMed: 9874779
- DOI: https://doi.org/10.1073/pnas.96.1.103
- Primary Citation of Related Structures:
9ILB - PubMed Abstract:
Water in the hydrophobic cavity of human interleukin 1beta, which was detected by NMR spectroscopy but was invisible by high resolution x-ray crystallography, has been mapped quantitatively by measurement and phasing of all of the low resolution x-ray diffraction data from a single crystal. Phases for the low resolution data were refined by iterative density modification of an initial flat solvent model outside the envelope of the atomic model. The refinement was restrained by the condition that the map of the difference between the electron density distribution in the full unit cell and that of the atomic model be flat within the envelope of the well ordered protein structure. Care was taken to avoid overfitting the diffraction data by maintaining phases for the high resolution data from the atomic model and by a resolution-dependent damping of the structure factor differences between data and model. The cavity region in the protein could accommodate up to four water molecules. The refined solvent difference map indicates that there are about two water molecules in the cavity region. This map is compatible with an atomic model of the water distribution refined by using XPLOR. About 70% of the time, there appears to be a water dimer in the central hydrophobic cavity, which is connected to the outside by two constricted channels occupied by single water molecules approximately 40% of the time on one side and approximately 10% on the other.
Organizational Affiliation:
Institute of Molecular Biophysics, Florida State University, Tallahassee, FL 32306-4380, USA.