Tricyclic sulfones as potent, selective and efficacious ROR gamma t inverse agonists - Exploring C6 and C8 SAR using late-stage functionalization.
Shi, Q., Xiao, Z., Yang, M.G., Marcoux, D., Cherney, R.J., Yip, S., Li, P., Wu, D.R., Weigelt, C.A., Sack, J., Khan, J., Ruzanov, M., Wang, J., Yarde, M., Ellen Cvijic, M., Li, S., Shuster, D.J., Xie, J., Sherry, T., Obermeier, M., Fura, A., Stefanski, K., Cornelius, G., Chacko, S., Shu, Y.Z., Khandelwal, P., Hynes Jr., J., Tino, J.A., Salter-Cid, L., Denton, R., Zhao, Q., Dhar, T.G.M.(2020) Bioorg Med Chem Lett 30: 127521-127521
- PubMed: 32882417
- DOI: https://doi.org/10.1016/j.bmcl.2020.127521
- Primary Citation of Related Structures:
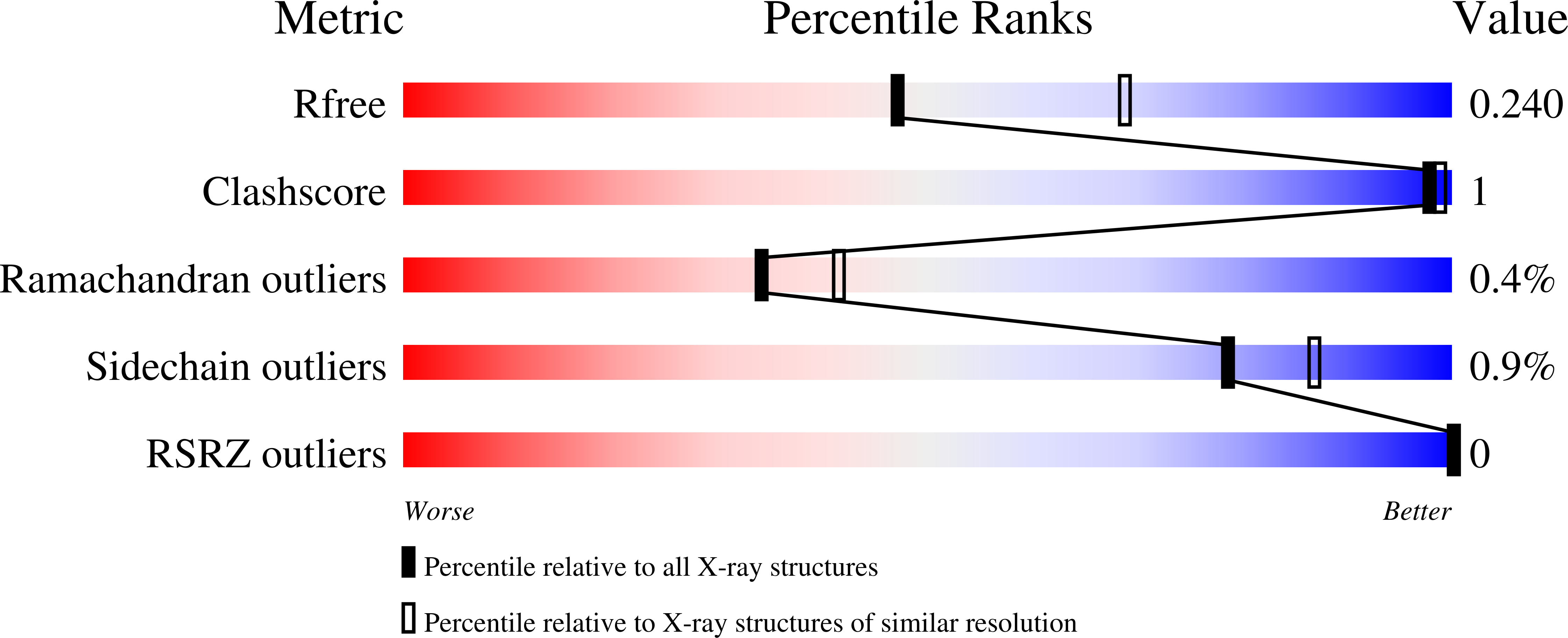
7JTM - PubMed Abstract:
In order to rapidly develop C6 and C8 SAR of our reported tricyclic sulfone series of RORγt inverse agonists, a late-stage bromination was employed. Although not regioselective, the bromination protocol allowed us to explore new substitution patterns/vectors that otherwise would have to be incorporated at the very beginning of the synthesis. Based on the SAR obtained from this exercise, compound 15 bearing a C8 fluorine was developed as a very potent and selective RORγt inverse agonist. This analog's in vitro profile, pharmacokinetic (PK) data and efficacy in an IL-23 induced mouse acanthosis model will be discussed.
Organizational Affiliation:
Research and Early Development, Bristol Myers Squibb, Route 206 and Province Line Road, Princeton, NJ 08543, United States. Electronic address: qing.shi@bms.com.