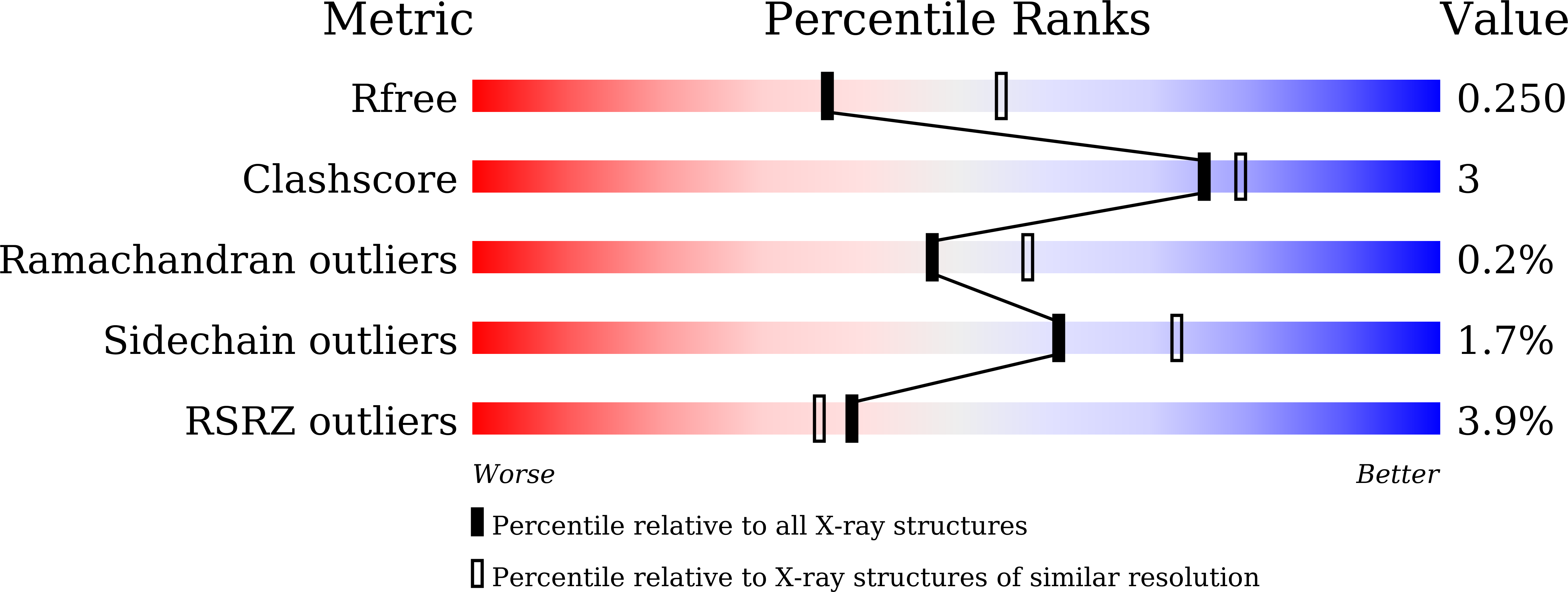
Structural Insights into the Regulation of Actin Capping Protein by Twinfilin C-terminal Tail.
Takeda, S., Koike, R., Fujiwara, I., Narita, A., Miyata, M., Ota, M., Maeda, Y.(2021) J Mol Biol 433: 166891-166891
- PubMed: 33639213
- DOI: https://doi.org/10.1016/j.jmb.2021.166891
- Primary Citation of Related Structures:
7DS2, 7DS3, 7DS4, 7DS6, 7DS8, 7DSA, 7DSB - PubMed Abstract:
Twinfilin is a conserved actin regulator that interacts with actin capping protein (CP) via C terminus residues (TWtail) that exhibits sequence similarity with the CP interaction (CPI) motif of CARMIL. Here we report the crystal structure of TWtail in complex with CP. Our structure showed that although TWtail and CARMIL CPI bind CP to an overlapping surface via their middle regions, they exhibit different CP-binding modes at both termini. Consequently, TWtail and CARMIL CPI restrict the CP in distinct conformations of open and closed forms, respectively. Interestingly, V-1, which targets CP away from the TWtail binding site, also favors the open-form CP. Consistently, TWtail forms a stable ternary complex with CP and V-1, a striking contrast to CARMIL CPI, which rapidly dissociates V-1 from CP. Our results demonstrate that TWtail is a unique CP-binding motif that regulates CP in a manner distinct from CARMIL CPI.
Organizational Affiliation:
Graduate School of Science, Nagoya University, Nagoya, Aichi 464-8602, Japan. Electronic address: takedashuichi@okayama-u.ac.jp.