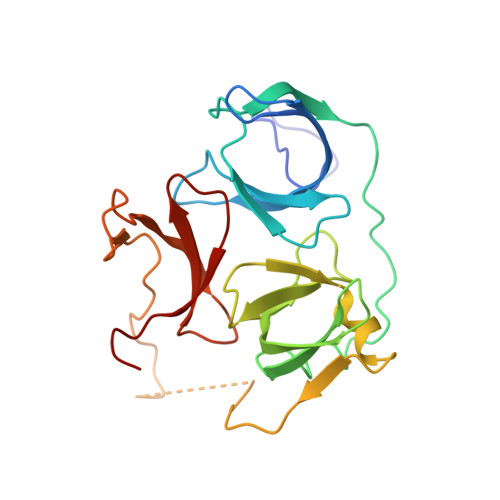
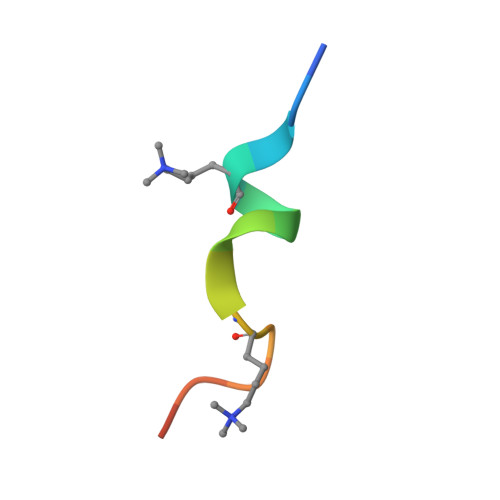
Molecular basis for histone H3 "K4me3-K9me3/2" methylation pattern readout by Spindlin1.
Zhao, F., Liu, Y., Su, X., Lee, J.E., Song, Y., Wang, D., Ge, K., Gao, J., Zhang, M.Q., Li, H.(2020) J Biol Chem 295: 16877-16887
- PubMed: 32994220
- DOI: https://doi.org/10.1074/jbc.RA120.013649
- Primary Citation of Related Structures:
7BQZ, 7BU9 - PubMed Abstract:
Histone recognition by "reader" modules serves as a fundamental mechanism in epigenetic regulation. Previous studies have shown that Spindlin1 is a reader of histone H3K4me3 as well as "K4me3-R8me2a" and promotes transcription of rDNA or Wnt/TCF4 target genes. Here we show that Spindlin1 also acts as a potent reader of histone H3 "K4me3-K9me3/2" bivalent methylation pattern. Calorimetric titration revealed a binding affinity of 16 nm between Spindlin1 and H3 "K4me3-K9me3" peptide, which is one to three orders of magnitude stronger than most other histone readout events at peptide level. Structural studies revealed concurrent recognition of H3K4me3 and H3K9me3/2 by aromatic pockets 2 and 1 of Spindlin1, respectively. Epigenomic profiling studies showed that Spindlin1 colocalizes with both H3K4me3 and H3K9me3 peaks in a subset of genes enriched in biological processes of transcription and its regulation. Moreover, the distribution of Spindlin1 peaks is primarily associated with H3K4me3 but not H3K9me3, which suggests that Spindlin1 is a downstream effector of H3K4me3 generated in heterochromatic regions. Collectively, our work calls attention to an intriguing function of Spindlin1 as a potent H3 "K4me3-K9me3/2" bivalent mark reader, thereby balancing gene expression and silencing in H3K9me3/2-enriched regions.
Organizational Affiliation:
MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing, China.