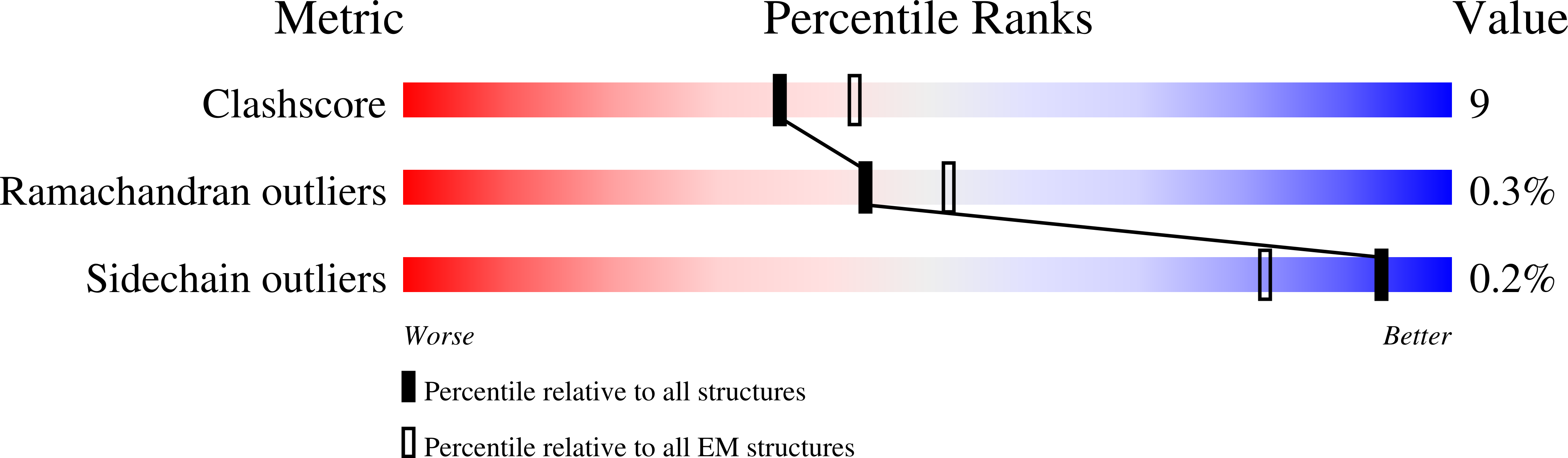Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Finogenova, K., Bonnet, J., Poepsel, S., Schafer, I.B., Finkl, K., Schmid, K., Litz, C., Strauss, M., Benda, C., Muller, J.(2020) Elife 9
- PubMed: 33211010
- DOI: https://doi.org/10.7554/eLife.61964
- Primary Citation of Related Structures:
7AT8 - PubMed Abstract:
Repression of genes by Polycomb requires that PRC2 modifies their chromatin by trimethylating lysine 27 on histone H3 (H3K27me3). At transcriptionally active genes, di- and tri-methylated H3K36 inhibit PRC2. Here, the cryo-EM structure of PRC2 on dinucleosomes reveals how binding of its catalytic subunit EZH2 to nucleosomal DNA orients the H3 N-terminus via an extended network of interactions to place H3K27 into the active site. Unmodified H3K36 occupies a critical position in the EZH2-DNA interface. Mutation of H3K36 to arginine or alanine inhibits H3K27 methylation by PRC2 on nucleosomes in vitro . Accordingly, Drosophila H3K36A and H3K36R mutants show reduced levels of H3K27me3 and defective Polycomb repression of HOX genes. The relay of interactions between EZH2, the nucleosomal DNA and the H3 N-terminus therefore creates the geometry that permits allosteric inhibition of PRC2 by methylated H3K36 in transcriptionally active chromatin.
Organizational Affiliation:
Max Planck Institute of Biochemistry, Laboratory of Chromatin Biology, Martinsried, Germany.