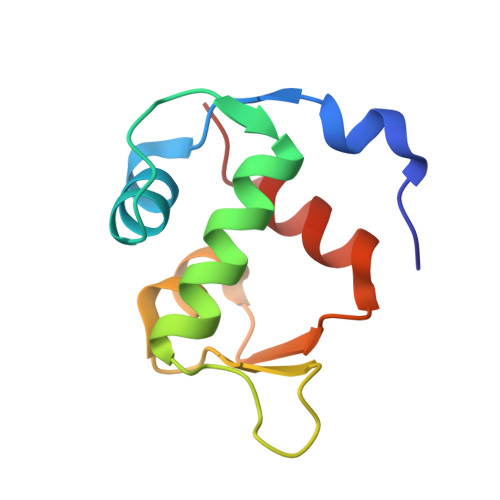
Diarylethene moiety as an enthalpy-entropy switch: photoisomerizable stapled peptides for modulating p53/MDM2 interaction.
Strizhak, A.V., Babii, O., Afonin, S., Bakanovich, I., Pantelejevs, T., Xu, W., Fowler, E., Eapen, R., Sharma, K., Platonov, M.O., Hurmach, V.V., Itzhaki, L., Hyvonen, M., Ulrich, A.S., Spring, D.R., Komarov, I.V.(2020) Org Biomol Chem 18: 5359-5369
- PubMed: 32390036
- DOI: https://doi.org/10.1039/d0ob00831a
- Primary Citation of Related Structures:
6Y4Q - PubMed Abstract:
Analogs of the known inhibitor (peptide pDI) of the p53/MDM2 protein-protein interaction are reported, which are stapled by linkers bearing a photoisomerizable diarylethene moiety. The corresponding photoisomers possess significantly different affinities to the p53-interacting domain of the human MDM2. Apparent dissociation constants are in the picomolar-to-low nanomolar range for those isomers with diarylethene in the "open" configuration, but up to eight times larger for the corresponding "closed" isomers. Spectroscopic, structural, and computational studies showed that the stapling linkers of the peptides contribute to their binding. Calorimetry revealed that the binding of the "closed" isomers is mostly enthalpy-driven, whereas the "open" photoforms bind to the protein stronger due to their increased binding entropy. The results suggest that conformational dynamics of the protein-peptide complexes may explain the differences in the thermodynamic profiles of the binding.
Organizational Affiliation:
University Chemical Laboratory, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK. Spring@ch.cam.ac.uk and Enamine Ltd, Vul. Chervonotkatska 78, 02094 Kyiv, Ukraine.