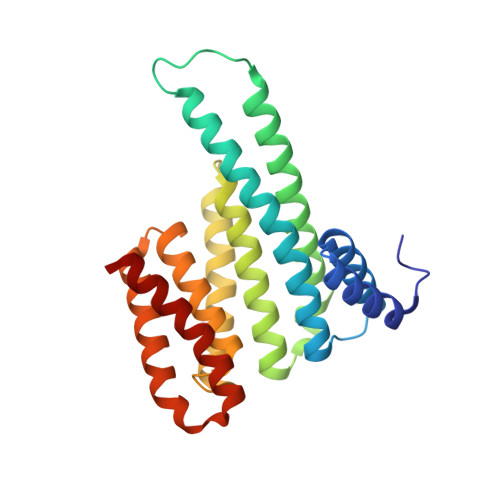
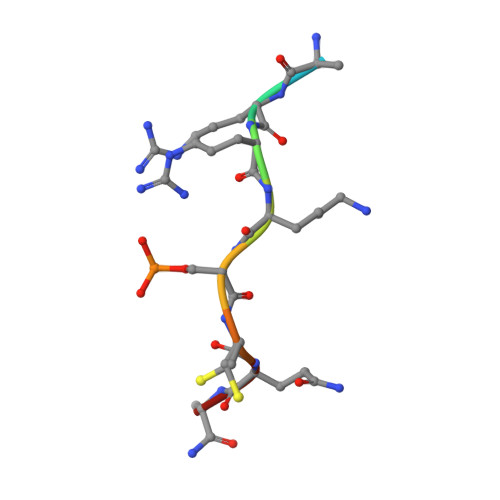
Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Sijbesma, E., Somsen, B.A., Miley, G.P., Leijten-van de Gevel, I.A., Brunsveld, L., Arkin, M.R., Ottmann, C.(2020) ACS Chem Biol 15: 3143-3148
- PubMed: 33196173
- DOI: https://doi.org/10.1021/acschembio.0c00646
- Primary Citation of Related Structures:
6XXC, 6XY5, 6Y18, 6Y1D, 6Y3W, 6Y58 - PubMed Abstract:
Protein-protein interaction (PPI) networks are fundamental for cellular processes. Small-molecule PPI enhancers have been shown to be powerful tools to fundamentally study PPIs and as starting points for potential new therapeutics. Yet, systematic approaches for their discovery are not widely available, and the design prerequisites of "molecular glues" are poorly understood. Covalent fragment-based screening can identify chemical starting points for these enhancers at specific sites in PPI interfaces. We recently reported a mass spectrometry-based disulfide-trapping (tethering) approach for a cysteine residue in the hub protein 14-3-3, an important regulator of phosphorylated client proteins. Here, we invert the strategy and report the development of a functional read-out for systematic identification of PPI enhancers based on fluorescence anisotropy (FA-tethering) with the reactive handle now on a client-derived peptide. Using the DNA-binding domain of the nuclear receptor Estrogen Related Receptor gamma (ERRγ), we target a native cysteine positioned at the 14-3-3 PPI interface and identify several fragments that form a disulfide bond to ERRγ and stabilize the complex up to 5-fold. Crystallography indicates that fragments bind in a pocket comprised of 14-3-3 and the ERRγ phosphopeptide. FA-tethering presents a streamlined methodology to discover molecular glues for protein complexes.
Organizational Affiliation:
Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, The Netherlands.