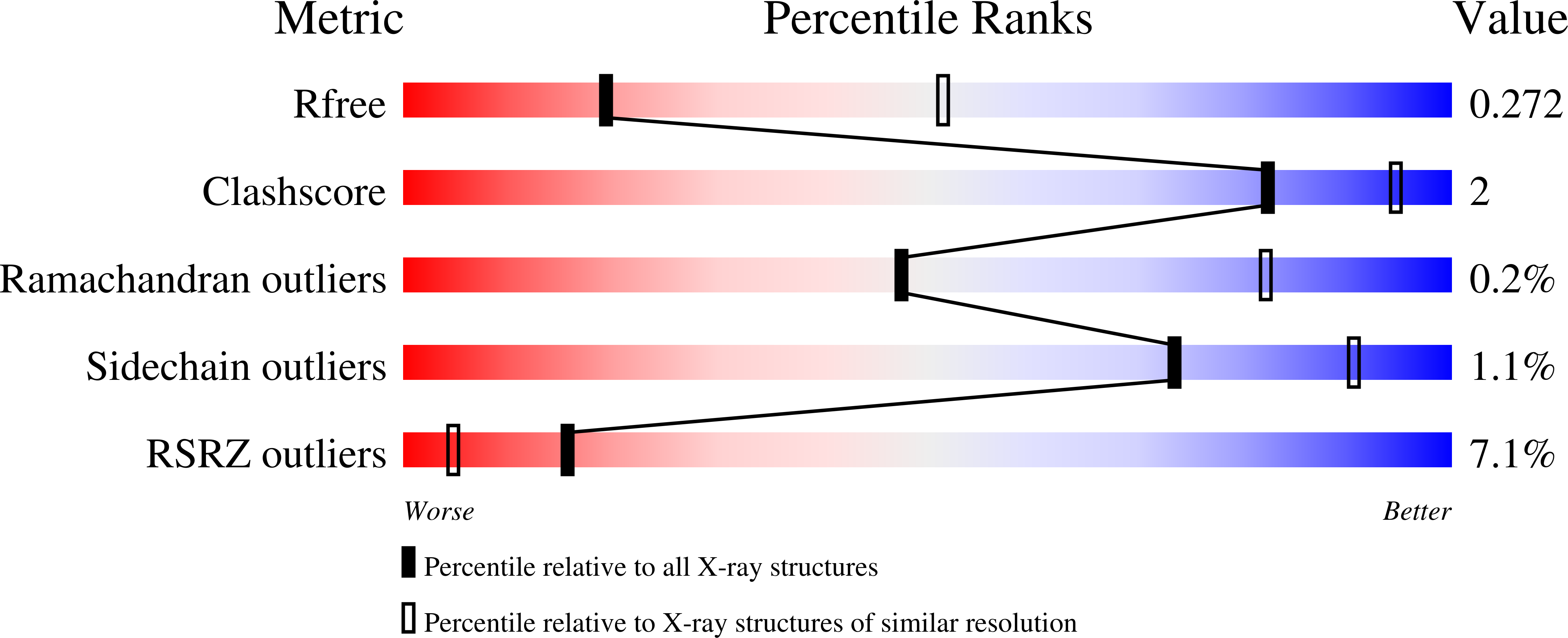
Discovery of Potent and Selective PI3K gamma Inhibitors.
Drew, S.L., Thomas-Tran, R., Beatty, J.W., Fournier, J., Lawson, K.V., Miles, D.H., Mata, G., Sharif, E.U., Yan, X., Mailyan, A.K., Ginn, E., Chen, J., Wong, K., Soni, D., Dhanota, P., Chen, P.Y., Shaqfeh, S.G., Meleza, C., Pham, A.T., Chen, A., Zhao, X., Banuelos, J., Jin, L., Schindler, U., Walters, M.J., Young, S.W., Walker, N.P., Leleti, M.R., Powers, J.P., Jeffrey, J.L.(2020) J Med Chem 63: 11235-11257
- PubMed: 32865410
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01203
- Primary Citation of Related Structures:
6XRL, 6XRM - PubMed Abstract:
The selective inhibition of the lipid signaling enzyme PI3Kγ constitutes an opportunity to mediate immunosuppression and inflammation within the tumor microenvironment but is difficult to achieve due to the high sequence homology across the class I PI3K isoforms. Here, we describe the design of a novel series of potent PI3Kγ inhibitors that attain high isoform selectivity through the divergent projection of substituents into both the "selectivity" and "alkyl-induced" pockets within the adenosine triphosphate (ATP) binding site of PI3Kγ. These efforts have culminated in the discovery of 5-[2-amino-3-(1-methyl-1 H -pyrazol-4-yl)pyrazolo[1,5- a ]pyrimidin-5-yl]-2-[(1 S )-1-cyclopropylethyl]-7-(trifluoromethyl)-2,3-dihydro-1 H -isoindol-1-one ( 4 , IC 50 = 0.064 μM, THP-1 cells), which displays >600-fold selectivity for PI3Kγ over the other class I isoforms and is a promising step toward the identification of a clinical development candidate. The structure-activity relationships identified throughout this campaign demonstrate that greater γ-selectivity can be achieved by inhibitors that occupy an "alkyl-induced" pocket and possess bicyclic hinge-binding motifs capable of forming more than one hydrogen bond to the hinge region of PI3Kγ.
Organizational Affiliation:
Arcus Biosciences, Inc., 3928 Point Eden Way, Hayward, California 94545, United States.