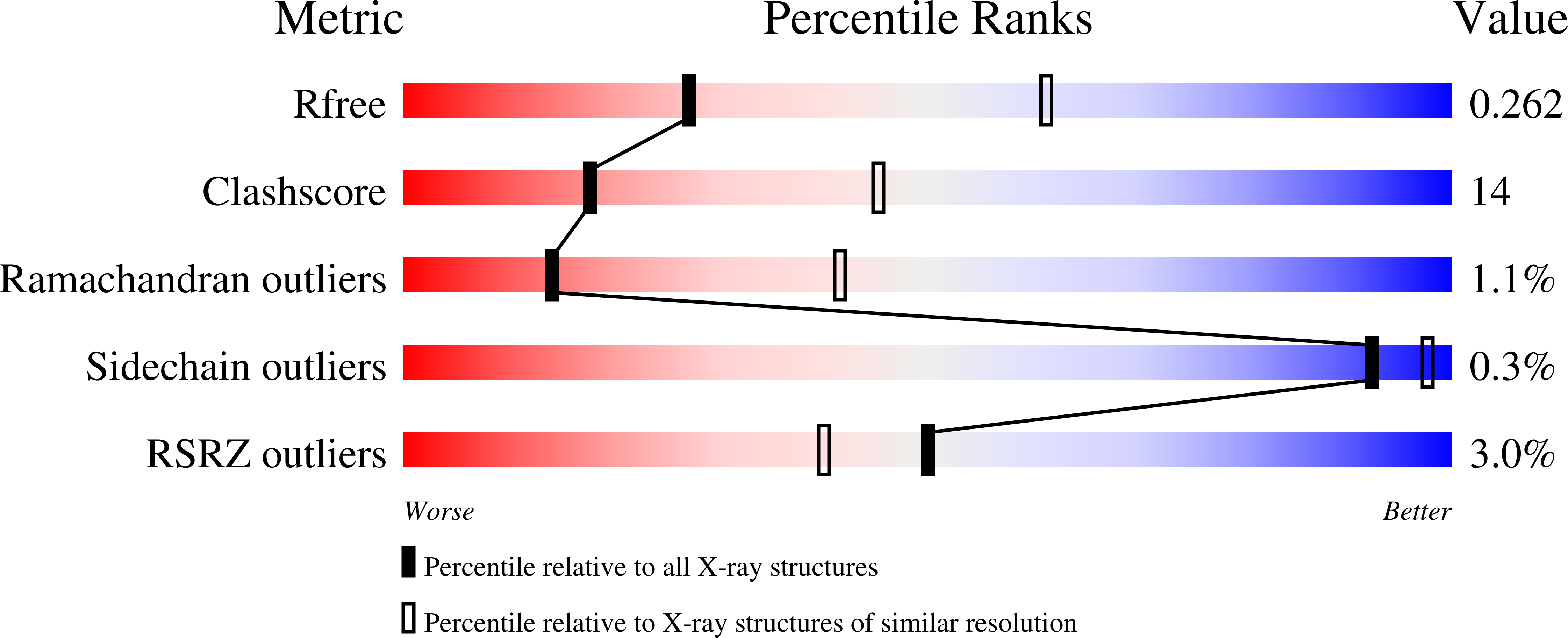
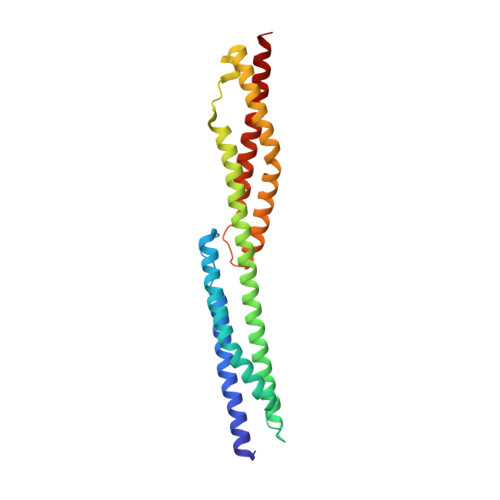
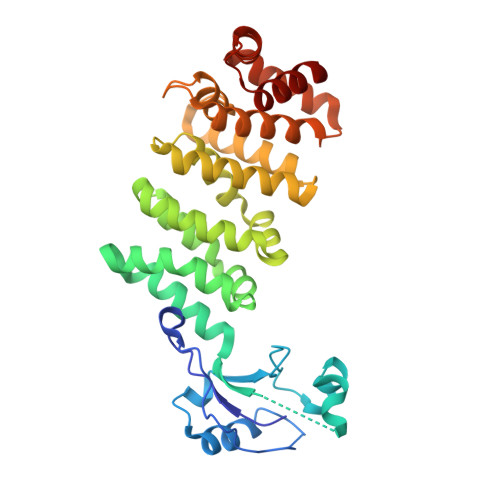
Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins.
Lim, S.M., Cruz, V.E., Antoku, S., Gundersen, G.G., Schwartz, T.U.(2021) Structure 29: 540-552.e5
- PubMed: 33472039
- DOI: https://doi.org/10.1016/j.str.2020.12.013
- Primary Citation of Related Structures:
6XF1, 6XF2 - PubMed Abstract:
The nuclear position in eukaryotes is controlled by a nucleo-cytoskeletal network, critical in cell differentiation, division, and movement. Forces are transmitted through conserved Linker of Nucleoskeleton and Cytoskeleton (LINC) complexes that traverse the nuclear envelope and engage on either side of the membrane with diverse binding partners. Nesprin-2-giant (Nes2G), a LINC element in the outer nuclear membrane, connects to the actin directly as well as through FHOD1, a formin primarily involved in actin bundling. Here, we report the crystal structure of Nes2G bound to FHOD1 and show that the presumed G-binding domain of FHOD1 is rather a spectrin repeat (SR) binding enhancer for the neighboring FH3 domain. The structure reveals that SR binding by FHOD1 is likely not regulated by the diaphanous-autoregulatory domain helix of FHOD1. Finally, we establish that Nes1G also has one FHOD1 binding SR, indicating that these abundant, giant Nesprins have overlapping functions in actin-bundle recruitment for nuclear movement.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.