Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Gaubitz, C., Liu, X., Magrino, J., Stone, N.P., Landeck, J., Hedglin, M., Kelch, B.A.(2020) bioRxiv
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2020) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication factor C subunit 1 | 595 | Homo sapiens | Mutation(s): 0 Gene Names: RFC1, RFC140 | 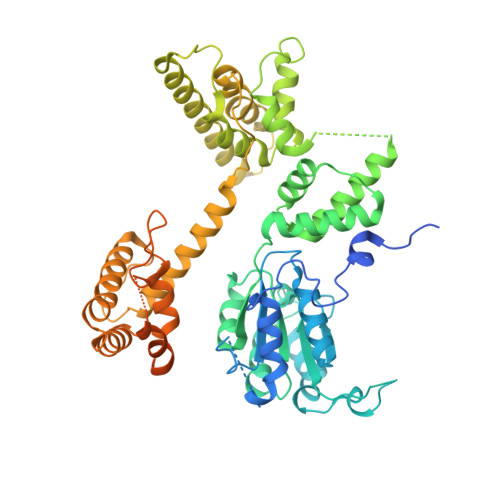 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P35251 (Homo sapiens) Explore P35251 Go to UniProtKB: P35251 | |||||
PHAROS: P35251 GTEx: ENSG00000035928 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35251 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication factor C subunit 2 | 354 | Homo sapiens | Mutation(s): 0 Gene Names: RFC2 | 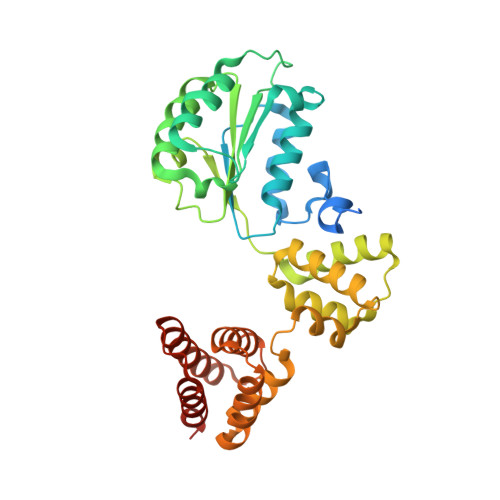 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P35250 (Homo sapiens) Explore P35250 Go to UniProtKB: P35250 | |||||
PHAROS: P35250 GTEx: ENSG00000049541 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35250 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication factor C subunit 5 | 340 | Homo sapiens | Mutation(s): 0 Gene Names: RFC5 | 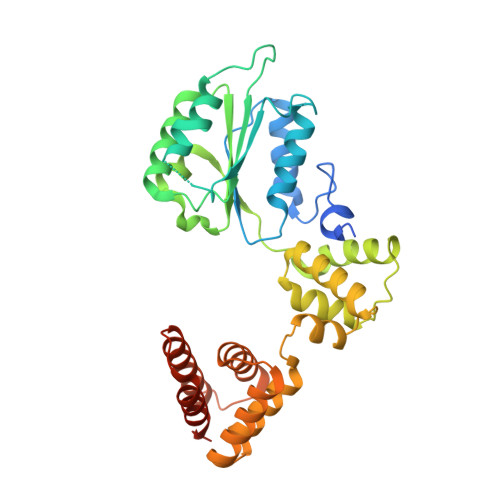 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P40937 (Homo sapiens) Explore P40937 Go to UniProtKB: P40937 | |||||
PHAROS: P40937 GTEx: ENSG00000111445 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P40937 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication factor C subunit 4 | 363 | Homo sapiens | Mutation(s): 0 Gene Names: RFC4 | 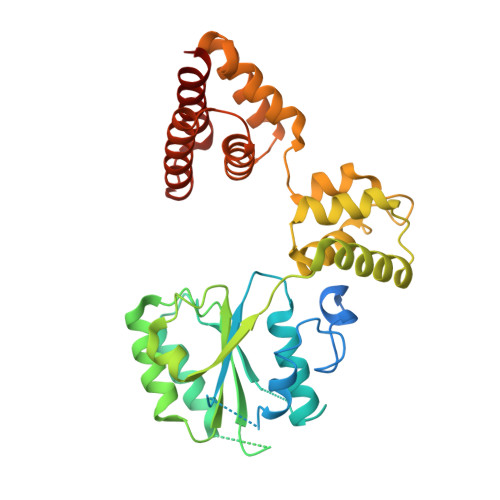 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P35249 (Homo sapiens) Explore P35249 Go to UniProtKB: P35249 | |||||
PHAROS: P35249 GTEx: ENSG00000163918 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35249 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication factor C subunit 3 | 356 | Homo sapiens | Mutation(s): 0 Gene Names: RFC3 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P40938 (Homo sapiens) Explore P40938 Go to UniProtKB: P40938 | |||||
PHAROS: P40938 GTEx: ENSG00000133119 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P40938 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Proliferating cell nuclear antigen | 261 | Homo sapiens | Mutation(s): 0 Gene Names: PCNA |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P12004 (Homo sapiens) Explore P12004 Go to UniProtKB: P12004 | |||||
PHAROS: P12004 GTEx: ENSG00000132646 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12004 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AGS (Subject of Investigation/LOI) Query on AGS | J [auth A], L [auth B], N [auth C], P [auth D] | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER C10 H16 N5 O12 P3 S NLTUCYMLOPLUHL-KQYNXXCUSA-N |  | ||
| ADP (Subject of Investigation/LOI) Query on ADP | Q [auth E] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | I [auth A], K [auth B], M [auth C], O [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | RELION | 3.0.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| American Cancer Society | United States | 440685 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R01-GM127776 |
| Swiss National Science Foundation | Switzerland | 177859 |