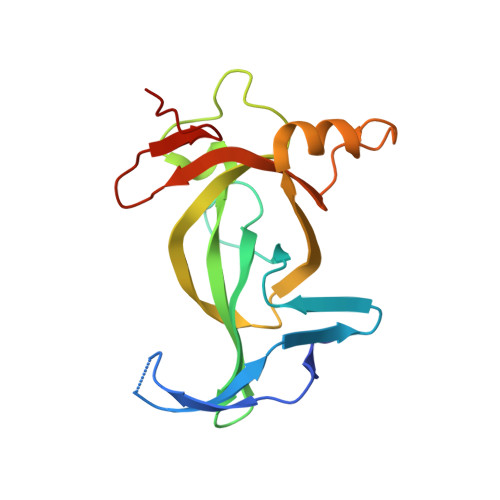
BAHCC1 binds H3K27me3 via a conserved BAH module to mediate gene silencing and oncogenesis.
Fan, H., Lu, J., Guo, Y., Li, D., Zhang, Z.M., Tsai, Y.H., Pi, W.C., Ahn, J.H., Gong, W., Xiang, Y., Allison, D.F., Geng, H., He, S., Diao, Y., Chen, W.Y., Strahl, B.D., Cai, L., Song, J., Wang, G.G.(2020) Nat Genet 52: 1384-1396
- PubMed: 33139953
- DOI: https://doi.org/10.1038/s41588-020-00729-3
- Primary Citation of Related Structures:
6VIL - PubMed Abstract:
Trimethylated histone H3 lysine 27 (H3K27me3) regulates gene repression, cell-fate determination and differentiation. We report that a conserved bromo-adjacent homology (BAH) module of BAHCC1 (BAHCC1 BAH ) 'recognizes' H3K27me3 specifically and enforces silencing of H3K27me3-demarcated genes in mammalian cells. Biochemical, structural and integrated chromatin immunoprecipitation-sequencing-based analyses demonstrate that direct readout of H3K27me3 by BAHCC1 is achieved through a hydrophobic trimethyl-L-lysine-binding 'cage' formed by BAHCC1 BAH , mediating colocalization of BAHCC1 and H3K27me3-marked genes. BAHCC1 is highly expressed in human acute leukemia and interacts with transcriptional corepressors. In leukemia, depletion of BAHCC1, or disruption of the BAHCC1 BAH -H3K27me3 interaction, causes derepression of H3K27me3-targeted genes that are involved in tumor suppression and cell differentiation, leading to suppression of oncogenesis. In mice, introduction of a germline mutation at Bahcc1 to disrupt its H3K27me3 engagement causes partial postnatal lethality, supporting a role in development. This study identifies an H3K27me3-directed transduction pathway in mammals that relies on a conserved BAH 'reader'.
Organizational Affiliation:
Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC, USA.