Progress in the Field of Aldehyde Dehydrogenase Inhibitors: Novel Imidazo[1,2- a ]pyridines against the 1A Family.
Quattrini, L., Gelardi, E.L.M., Petrarolo, G., Colombo, G., Ferraris, D.M., Picarazzi, F., Rizzi, M., Garavaglia, S., La Motta, C.(2020) ACS Med Chem Lett 11: 963-970
- PubMed: 32435412
- DOI: https://doi.org/10.1021/acsmedchemlett.9b00686
- Primary Citation of Related Structures:
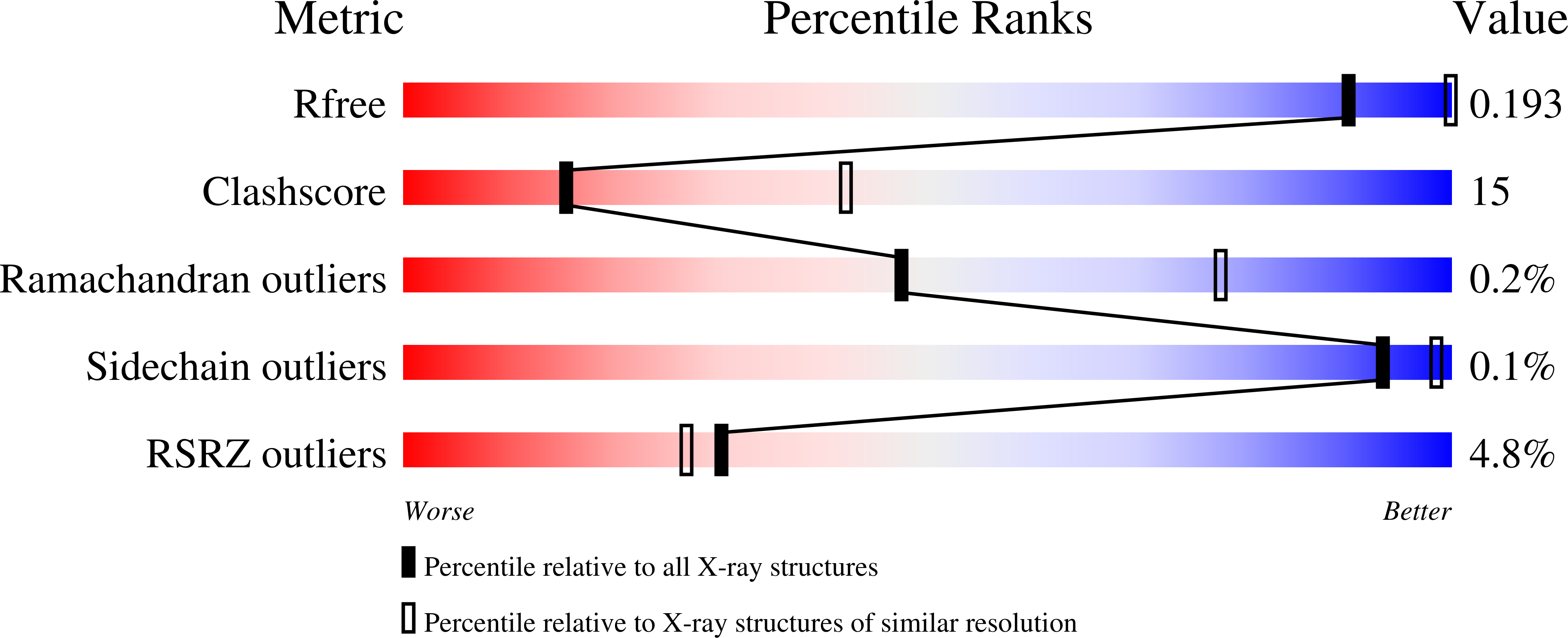
6TRY - PubMed Abstract:
Members of the aldehyde dehydrogenase 1A family are commonly acknowledged as hallmarks of cancer stem cells, and their overexpression is significantly associated with poor prognosis in different types of malignancies. Accordingly, treatments targeting these enzymes may represent a successful strategy to fight cancer. In this work we describe a novel series of imidazo[1,2- a ]pyridines, designed as aldehyde dehydrogenase inhibitors by means of a structure-based optimization of a previously developed lead. The novel compounds were evaluated in vitro for their activity and selectivity against the three isoforms of the ALDH1A family and investigated through crystallization and modeling studies for their ability to interact with the catalytic site of the 1A3 isoform. Compound 3f emerged as the first in class submicromolar competitive inhibitor of the target enzyme.
Organizational Affiliation:
Department of Pharmacy, University of Pisa, Via Bonanno 6, 56126 Pisa, Italy.