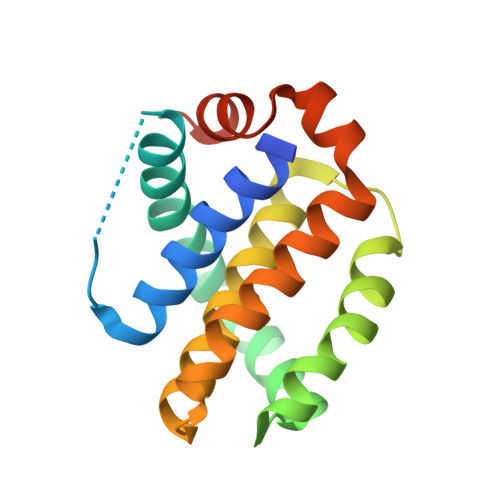
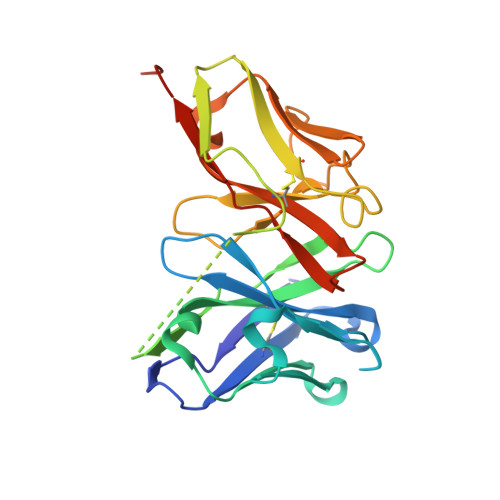
Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Luptak, J., Bista, M., Fisher, D., Flavell, L., Gao, N., Wickson, K., Kazmirski, S.L., Howard, T., Rawlins, P.B., Hargreaves, D.(2019) Acta Crystallogr D Struct Biol 75: 1003-1014
- PubMed: 31692474
- DOI: https://doi.org/10.1107/S2059798319014116
- Primary Citation of Related Structures:
6QB3, 6QB4, 6QB6, 6QB9, 6QBC, 6QF9, 6QFC - PubMed Abstract:
Apoptosis is a crucial process by which multicellular organisms control tissue growth, removal and inflammation. Disruption of the normal apoptotic function is often observed in cancer, where cell death is avoided by the overexpression of anti-apoptotic proteins of the Bcl-2 (B-cell lymphoma 2) family, including Mcl-1 (myeloid cell leukaemia 1). This makes Mcl-1 a potential target for drug therapy, through which normal apoptosis may be restored by inhibiting the protective function of Mcl-1. Here, the discovery and biophysical properties of an anti-Mcl-1 antibody fragment are described and the utility of both the scFv and Fab are demonstrated in generating an Mcl-1 crystal system amenable to iterative structure-guided drug design.
Organizational Affiliation:
Discovery Sciences, R&D Biopharmaceuticals, AstraZeneca, Cambridge CB4 0WG, England.