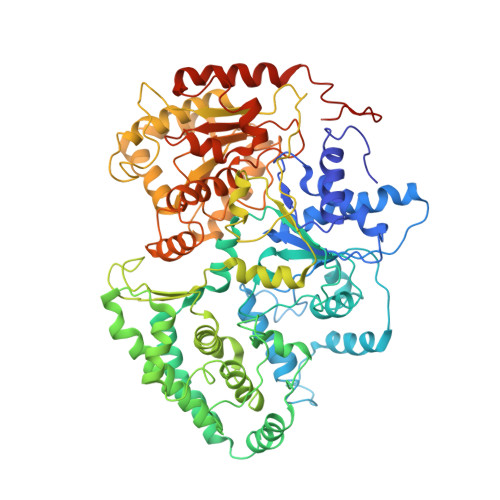
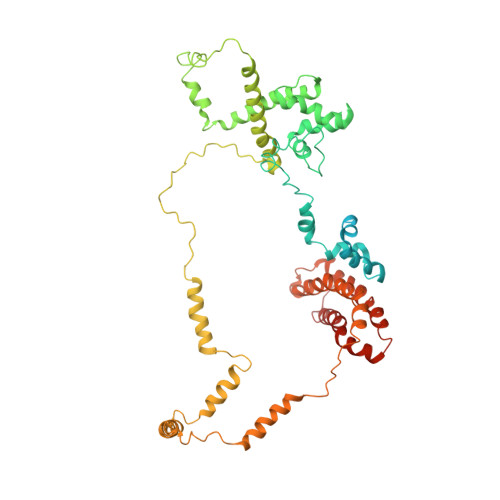
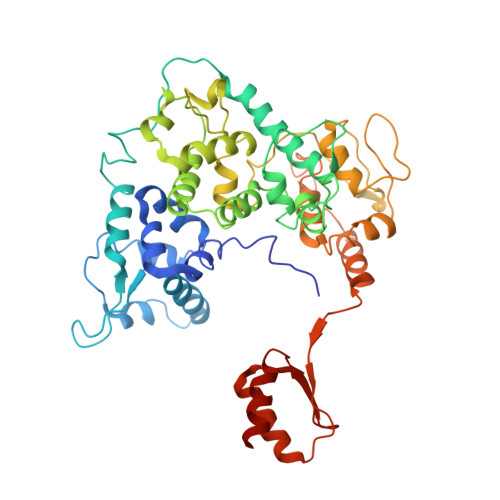
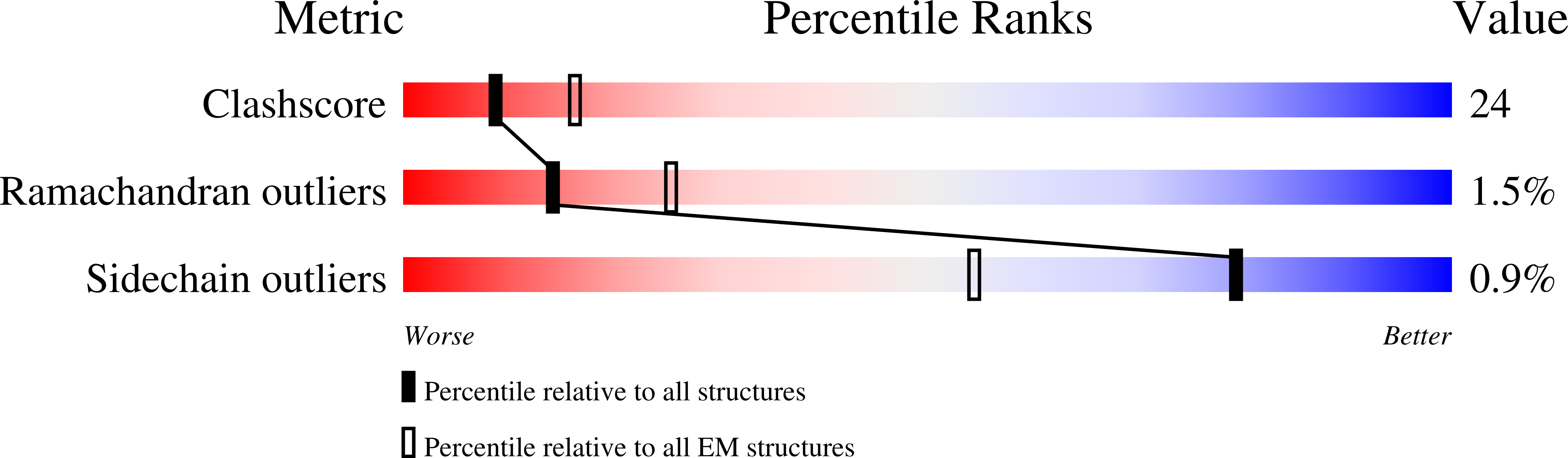Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Yan, C., Dodd, T., He, Y., Tainer, J.A., Tsutakawa, S.E., Ivanov, I.(2019) Nat Struct Mol Biol 26: 397-406
- PubMed: 31110295
- DOI: https://doi.org/10.1038/s41594-019-0220-3
- Primary Citation of Related Structures:
6O9L, 6O9M - PubMed Abstract:
Transcription preinitiation complexes (PICs) are vital assemblies whose function underlies the expression of protein-encoding genes. Cryo-EM advances have begun to uncover their structural organization. Nevertheless, functional analyses are hindered by incompletely modeled regions. Here we integrate all available cryo-EM data to build a practically complete human PIC structural model. This enables simulations that reveal the assembly's global motions, define PIC partitioning into dynamic communities and delineate how structural modules function together to remodel DNA. We identify key TFIIE-p62 interactions that link core-PIC to TFIIH. p62 rigging interlaces p34, p44 and XPD while capping the DNA-binding and ATP-binding sites of XPD. PIC kinks and locks substrate DNA, creating negative supercoiling within the Pol II cleft to facilitate promoter opening. Mapping disease mutations associated with xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome onto defined communities reveals clustering into three mechanistic classes that affect TFIIH helicase functions, protein interactions and interface dynamics.
Organizational Affiliation:
Department of Chemistry, Georgia State University, Atlanta, GA, USA.