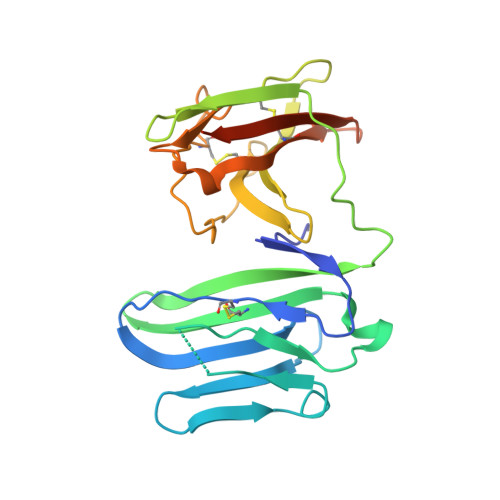
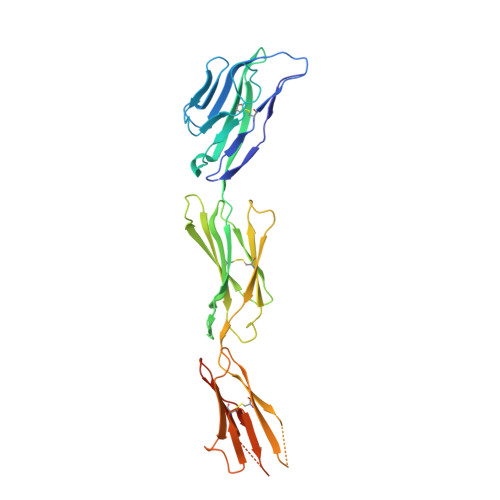
Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1.
Deuss, F.A., Watson, G.M., Goodall, K.J., Leece, I., Chatterjee, S., Fu, Z., Thaysen-Andersen, M., Andrews, D.M., Rossjohn, J., Berry, R.(2019) J Biol Chem 294: 12534-12546
- PubMed: 31253644
- DOI: https://doi.org/10.1074/jbc.RA119.009261
- Primary Citation of Related Structures:
6O3O - PubMed Abstract:
Nectin and nectin-like (Necl) adhesion molecules are broadly overexpressed in a wide range of cancers. By binding to these adhesion molecules, the immunoreceptors DNAX accessory molecule-1 (DNAM-1), CD96 molecule (CD96), and T-cell immunoreceptor with Ig and ITIM domains (TIGIT) play a crucial role in regulating the anticancer activities of immune effector cells. However, within this axis, it remains unclear how DNAM-1 recognizes its cognate ligands. Here, we determined the structure of human DNAM-1 in complex with nectin-like protein-5 (Necl-5) at 2.8 Å resolution. Unexpectedly, we found that the two extracellular domains (D1-D2) of DNAM-1 adopt an unconventional "collapsed" arrangement that is markedly distinct from those in other immunoglobulin-based immunoreceptors. The DNAM-1/Necl-5 interaction was underpinned by conserved lock-and-key motifs located within their respective D1 domains, but also included a distinct interface derived from DNAM-1 D2. Mutation of the signature DNAM-1 "key" motif within the D1 domain attenuated Necl-5 binding and natural killer cell-mediated cytotoxicity. Altogether, our results have implications for understanding the binding mode of an immune receptor family that is emerging as a viable candidate for cancer immunotherapy.
Organizational Affiliation:
Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria 3800, Australia.