RHODOCETIN IN COMPLEX WITH THE INTEGRIN ALPHA2-A DOMAIN WITH MANGANESE BOUND
Stetefeld, J., McDougall, M.D., Loewen, P.C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Snaclec rhodocetin subunit gamma | 135 | Calloselasma rhodostoma | Mutation(s): 0 | 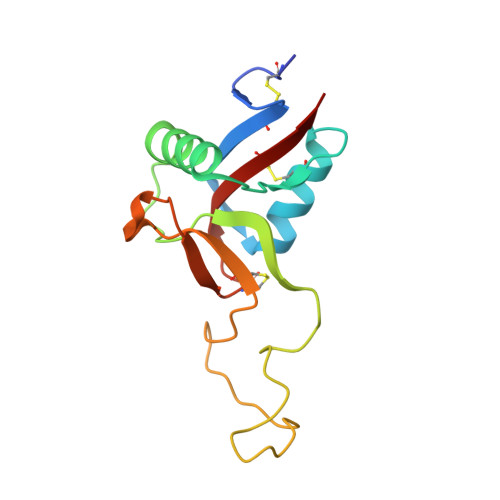 | |
UniProt | |||||
Find proteins for D2YW39 (Calloselasma rhodostoma) Explore D2YW39 Go to UniProtKB: D2YW39 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D2YW39 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Snaclec rhodocetin subunit delta | 124 | Calloselasma rhodostoma | Mutation(s): 0 | 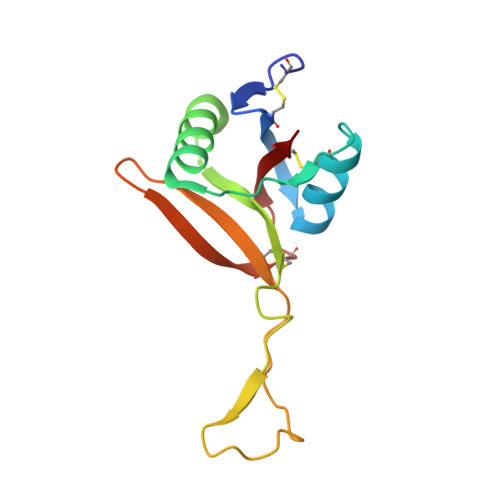 | |
UniProt | |||||
Find proteins for D2YW40 (Calloselasma rhodostoma) Explore D2YW40 Go to UniProtKB: D2YW40 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D2YW40 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Integrin alpha-2 | 217 | Homo sapiens | Mutation(s): 0 Gene Names: ITGA2, CD49B | 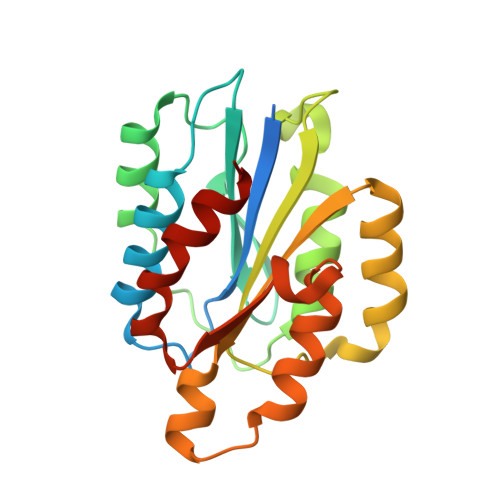 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P17301 (Homo sapiens) Explore P17301 Go to UniProtKB: P17301 | |||||
PHAROS: P17301 GTEx: ENSG00000164171 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P17301 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | AA [auth E] AB [auth R] DA [auth F] GA [auth G] JA [auth I] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| MN Query on MN | CA [auth F] IA [auth I] PA [auth L] U [auth C] UA [auth O] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| CL Query on CL | BA [auth E] BB [auth R] FA [auth G] HA [auth H] MA [auth J] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | CB [auth R] EA [auth F] KA [auth I] QA [auth L] VA [auth O] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| NH4 Query on NH4 | Z [auth D] | AMMONIUM ION H4 N QGZKDVFQNNGYKY-UHFFFAOYSA-O |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 130.712 | α = 90 |
| b = 130.712 | β = 90 |
| c = 251.8 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALA | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| REFMAC | phasing |