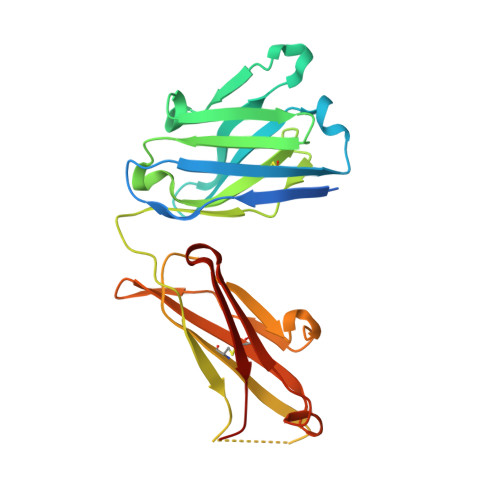
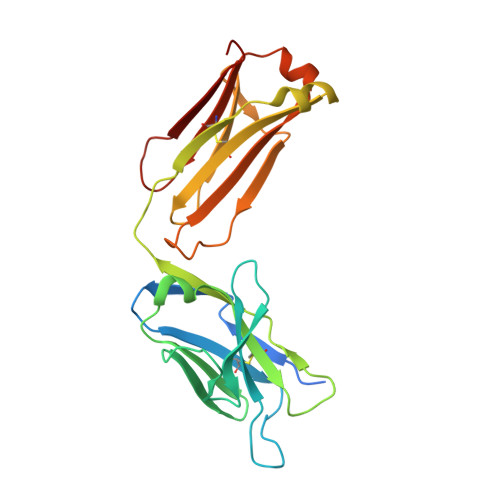
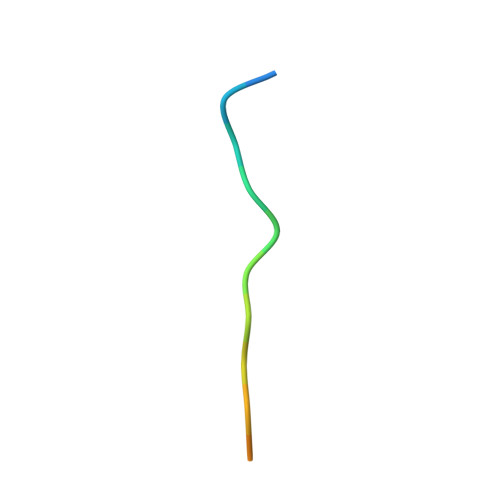
A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Wakui, H., Tanaka, Y., Ose, T., Matsumoto, I., Kato, K., Min, Y., Tachibana, T., Sato, M., Naruchi, K., Martin, F.G., Hinou, H., Nishimura, S.-I.(2020) Chem Sci 11: 4999-5006