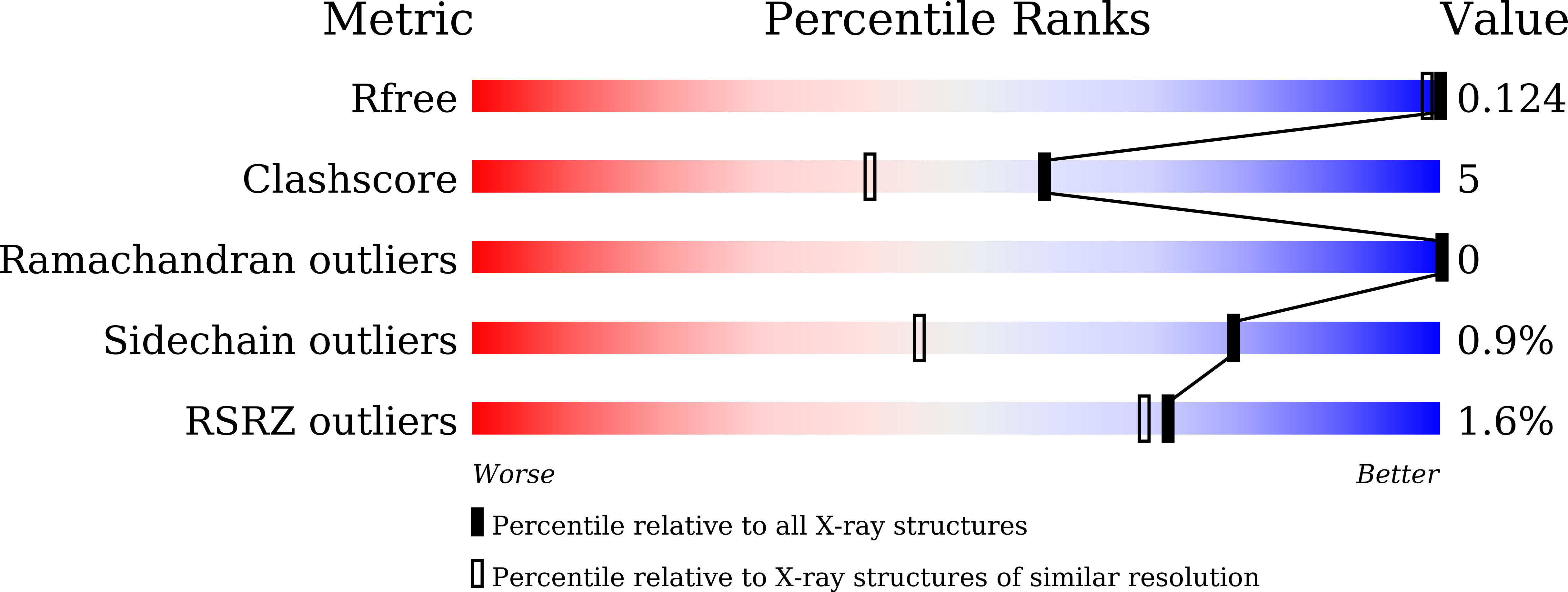
Structural insights into the effect of active-site mutation on the catalytic mechanism of carbonic anhydrase.
Kim, J.K., Lee, C., Lim, S.W., Andring, J.T., Adhikari, A., McKenna, R., Kim, C.U.(2020) IUCrJ 7: 985-994
- PubMed: 33209313
- DOI: https://doi.org/10.1107/S2052252520011008
- Primary Citation of Related Structures:
6KLZ, 6KM0, 6KM1, 6KM2, 6KM3, 6KM4, 6KM5, 6KM6 - PubMed Abstract:
Enzymes are catalysts of biological processes. Significant insight into their catalytic mechanisms has been obtained by relating site-directed mutagenesis studies to kinetic activity assays. However, revealing the detailed relationship between structural modifications and functional changes remains challenging owing to the lack of information on reaction intermediates and of a systematic way of connecting them to the measured kinetic parameters. Here, a systematic approach to investigate the effect of an active-site-residue mutation on a model enzyme, human carbonic anhydrase II (CA II), is described. Firstly, structural analysis is performed on the crystallographic intermediate states of native CA II and its V143I variant. The structural comparison shows that the binding affinities and configurations of the substrate (CO 2 ) and product (HCO 3 - ) are altered in the V143I variant and the water network in the water-replenishment pathway is restructured, while the proton-transfer pathway remains mostly unaffected. This structural information is then used to estimate the modifications of the reaction rate constants and the corresponding free-energy profiles of CA II catalysis. Finally, the obtained results are used to reveal the effect of the V143I mutation on the measured kinetic parameters ( k cat and k cat / K m ) at the atomic level. It is believed that the systematic approach outlined in this study may be used as a template to unravel the structure-function relationships of many other biologically important enzymes.
Organizational Affiliation:
Department of Physics, Ulsan National Institute of Science and Technology (UNIST), Ulsan 44919, Republic of Korea.