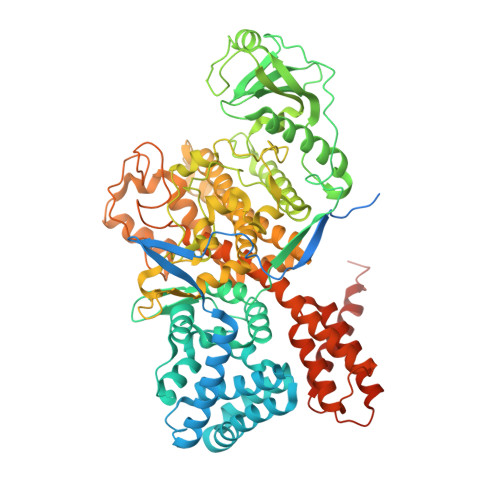
Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase.
Gan, N., Zhen, X., Liu, Y., Xu, X., He, C., Qiu, J., Liu, Y., Fujimoto, G.M., Nakayasu, E.S., Zhou, B., Zhao, L., Puvar, K., Das, C., Ouyang, S., Luo, Z.Q.(2019) Nature 572: 387-391
- PubMed: 31330531
- DOI: https://doi.org/10.1038/s41586-019-1439-1
- Primary Citation of Related Structures:
6K4K, 6K4L, 6K4R - PubMed Abstract:
The bacterial pathogen Legionella pneumophila creates an intracellular niche permissive for its replication by extensively modulating host-cell functions using hundreds of effector proteins delivered by its Dot/Icm secretion system 1 . Among these, members of the SidE family (SidEs) regulate several cellular processes through a unique phosphoribosyl ubiquitination mechanism that bypasses the canonical ubiquitination machinery 2-4 . The activity of SidEs is regulated by another Dot/Icm effector known as SidJ 5 ; however, the mechanism of this regulation is not completely understood 6,7 . Here we demonstrate that SidJ inhibits the activity of SidEs by inducing the covalent attachment of glutamate moieties to SdeA-a member of the SidE family-at E860, one of the catalytic residues that is required for the mono-ADP-ribosyltransferase activity involved in ubiquitin activation 2 . This inhibition by SidJ is spatially restricted in host cells because its activity requires the eukaryote-specific protein calmodulin (CaM). We solved a structure of SidJ-CaM in complex with AMP and found that the ATP used in this reaction is cleaved at the α-phosphate position by SidJ, which-in the absence of glutamate or modifiable SdeA-undergoes self-AMPylation. Our results reveal a mechanism of regulation in bacterial pathogenicity in which a glutamylation reaction that inhibits the activity of virulence factors is activated by host-factor-dependent acyl-adenylation.
Organizational Affiliation:
Purdue Institute for Inflammation, Immunology and Infectious Disease and Department of Biological Sciences, Purdue University, West Lafayette, IN, USA.