Structural diversity in the atomic resolution 3D fingerprint of the titin M-band segment.
Chatziefthimiou, S.D., Hornburg, P., Sauer, F., Mueller, S., Ugurlar, D., Xu, E.R., Wilmanns, M.(2019) PLoS One 14: e0226693-e0226693
- PubMed: 31856237
- DOI: https://doi.org/10.1371/journal.pone.0226693
- Primary Citation of Related Structures:
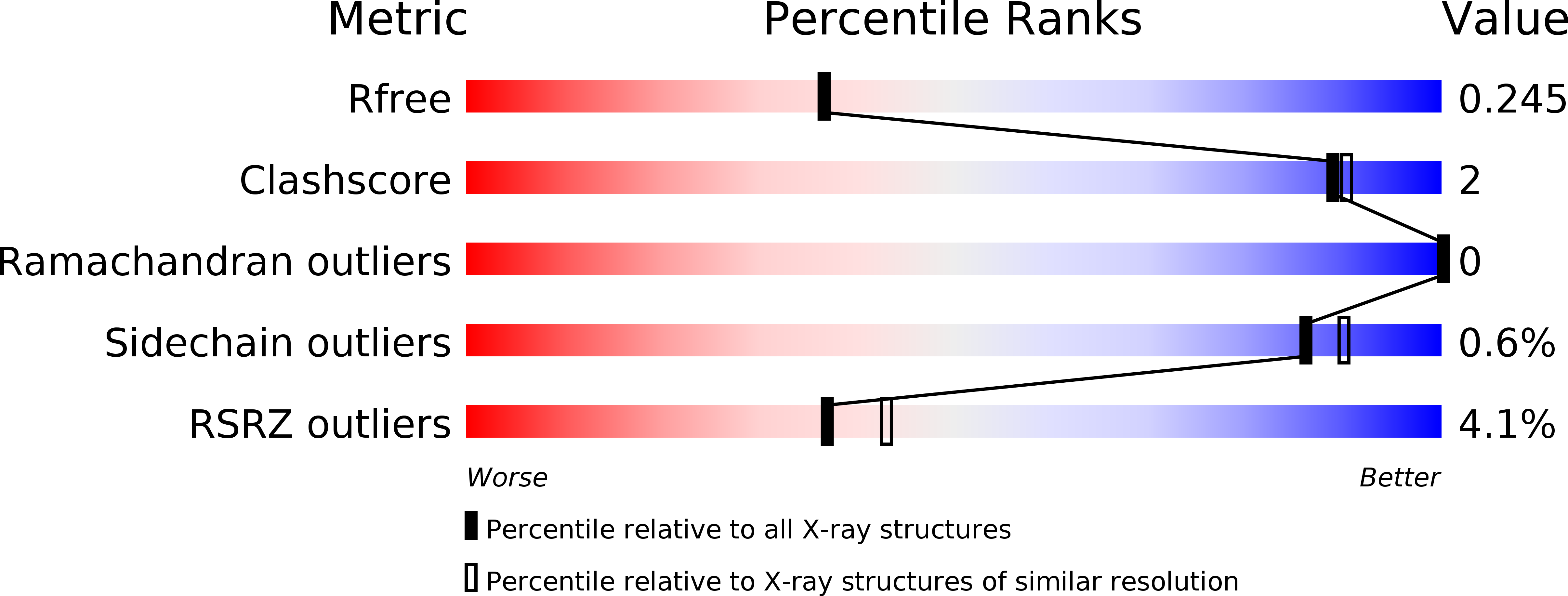
6H4L, 6HCI - PubMed Abstract:
In striated muscles, molecular filaments are largely composed of long protein chains with extensive arrays of identically folded domains, referred to as "beads-on-a-string". It remains a largely unresolved question how these domains have developed a unique molecular profile such that each carries out a distinct function without false-positive readout. This study focuses on the M-band segment of the sarcomeric protein titin, which comprises ten identically folded immunoglobulin domains. Comparative analysis of high-resolution structures of six of these domains ‒ M1, M3, M4, M5, M7, and M10 ‒ reveals considerable structural diversity within three distinct loops and a non-conserved pattern of exposed cysteines. Our data allow to structurally interpreting distinct pathological readouts that result from titinopathy-associated variants. Our findings support general principles that could be used to identify individual structural/functional profiles of hundreds of identically folded protein domains within the sarcomere and other densely crowded cellular environments.
Organizational Affiliation:
European Molecular Biology Laboratory, Hamburg Unit, Hamburg, Germany.