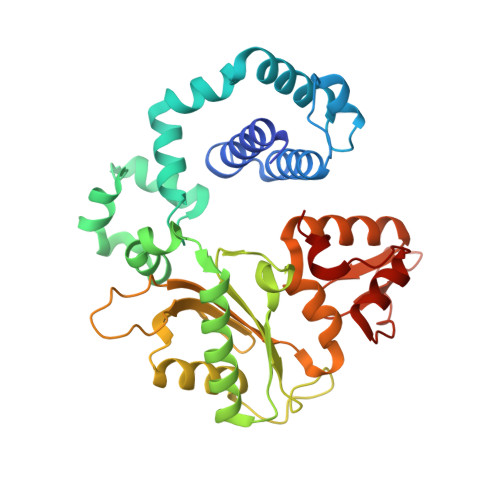
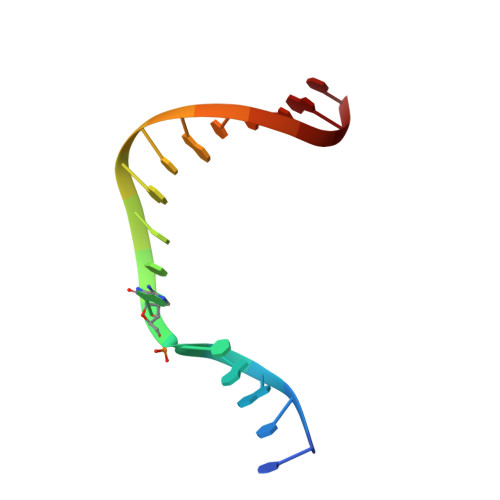
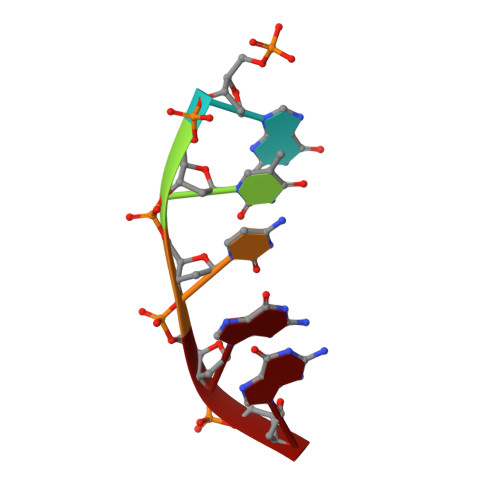
Mutagenic Replication of the Major Oxidative Adenine Lesion 7,8-Dihydro-8-oxoadenine by Human DNA Polymerases.
Koag, M.C., Jung, H., Lee, S.(2019) J Am Chem Soc 141: 4584-4596
- PubMed: 30817143
- DOI: https://doi.org/10.1021/jacs.8b08551
- Primary Citation of Related Structures:
6E3R, 6E3V, 6E3W - PubMed Abstract:
Reactive oxygen species attack DNA to produce 7,8-dihyro-8-oxoguanine (oxoG) and 7,8-dihydro-8-oxoadenine (oxoA) as major lesions. The structural basis for the mutagenicity of oxoG, which induces G to T mutations, is well understood. However, the structural basis for the mutagenic potential of oxoA, which induces A to C mutations, remains poorly understood. To gain insight into oxoA-induced mutagenesis, we conducted kinetic studies of human DNA polymerases β and η replicating across oxoA and structural studies of polβ incorporating dTTP/dGTP opposite oxoA. While polη readily bypassed oxoA, it incorporated dGTP opposite oxoA with a catalytic specificity comparable to that of correct insertion, underscoring the promutagenic nature of the major oxidative adenine lesion. Polη and polβ incorporated dGTP opposite oxoA ∼170-fold and ∼100-fold more efficiently than that opposite dA, respectively, indicating that the 8-oxo moiety greatly facilitated error-prone replication. Crystal structures of polβ showed that, when paired with an incoming dTTP, the templating oxoA adopted an anti conformation and formed Watson-Crick base pair. When paired with dGTP, oxoA adopted a syn conformation and formed a Hoogsteen base pair with Watson-Crick-like geometry, highlighting the dual-coding potential of oxoA. The templating oxoA was stabilized by Lys280-mediated stacking and hydrogen bonds. Overall, these results provide insight into the mutagenic potential and dual-coding nature of the major oxidative adenine lesion.
Organizational Affiliation:
The Division of Chemical Biology and Medicinal Chemistry, College of Pharmacy , The University of Texas at Austin , Austin , Texas 78712 , United States.