Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
Tumbale, P., Schellenberg, M.J., Mueller, G.A., Fairweather, E., Watson, M., Little, J.N., Krahn, J., Waddell, I., London, R.E., Williams, R.S.(2018) EMBO J 37
- PubMed: 29934293
- DOI: https://doi.org/10.15252/embj.201798875
- Primary Citation of Related Structures:
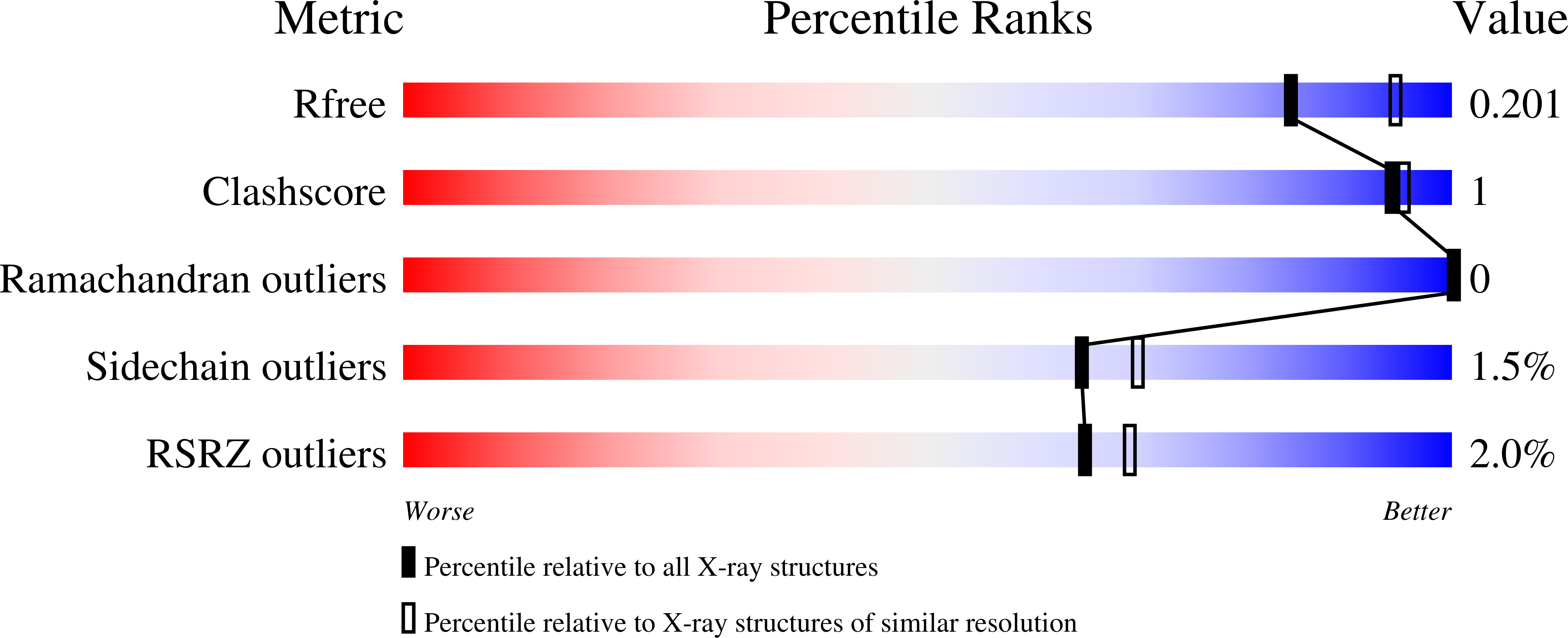
6CVO, 6CVP, 6CVQ, 6CVR, 6CVS, 6CVT - PubMed Abstract:
The failure of DNA ligases to complete their catalytic reactions generates cytotoxic adenylated DNA strand breaks. The APTX RNA-DNA deadenylase protects genome integrity and corrects abortive DNA ligation arising during ribonucleotide excision repair and base excision DNA repair, and APTX human mutations cause the neurodegenerative disorder ataxia with oculomotor ataxia 1 (AOA1). How APTX senses cognate DNA nicks and is inactivated in AOA1 remains incompletely defined. Here, we report X-ray structures of APTX engaging nicked RNA-DNA substrates that provide direct evidence for a wedge-pivot-cut strategy for 5'-AMP resolution shared with the alternate 5'-AMP processing enzymes POLβ and FEN1. Our results uncover a DNA-induced fit mechanism regulating APTX active site loop conformations and assembly of a catalytically competent active center. Further, based on comprehensive biochemical, X-ray and solution NMR results, we define a complex hierarchy for the differential impacts of the AOA1 mutational spectrum on APTX structure and activity. Sixteen AOA1 variants impact APTX protein stability, one mutation directly alters deadenylation reaction chemistry, and a dominant AOA1 variant unexpectedly allosterically modulates APTX active site conformations.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, Department of Health and Human Services, National Institute of Environmental Health Sciences, US National Institutes of Health, Research Triangle Park, NC, USA.