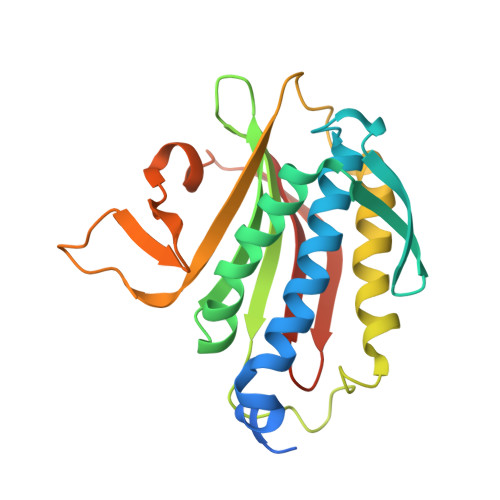
Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Rizzo, A.A., Vassel, F.M., Chatterjee, N., D'Souza, S., Li, Y., Hao, B., Hemann, M.T., Walker, G.C., Korzhnev, D.M.(2018) Proc Natl Acad Sci U S A 115: E8191-E8200
- PubMed: 30111544
- DOI: https://doi.org/10.1073/pnas.1801149115
- Primary Citation of Related Structures:
6BC8, 6BCD, 6BI7 - PubMed Abstract:
The translesion synthesis (TLS) polymerases Polζ and Rev1 form a complex that enables replication of damaged DNA. The Rev7 subunit of Polζ, which is a multifaceted HORMA (Hop1, Rev7, Mad2) protein with roles in TLS, DNA repair, and cell-cycle control, facilitates assembly of this complex by binding Rev1 and the catalytic subunit of Polζ, Rev3. Rev7 interacts with Rev3 by a mechanism conserved among HORMA proteins, whereby an open-to-closed transition locks the ligand underneath the "safety belt" loop. Dimerization of HORMA proteins promotes binding and release of this ligand, as exemplified by the Rev7 homolog, Mad2. Here, we investigate the dimerization of Rev7 when bound to the two Rev7-binding motifs (RBMs) in Rev3 by combining in vitro analyses of Rev7 structure and interactions with a functional assay in a Rev7 -/- cell line. We demonstrate that Rev7 uses the conventional HORMA dimerization interface both to form a homodimer when tethered by the two RBMs in Rev3 and to heterodimerize with other HORMA domains, Mad2 and p31 comet Structurally, the Rev7 dimer can bind only one copy of Rev1, revealing an unexpected Rev1/Polζ architecture. In cells, mutation of the Rev7 dimer interface increases sensitivity to DNA damage. These results provide insights into the structure of the Rev1/Polζ TLS assembly and highlight the function of Rev7 homo- and heterodimerization.
Organizational Affiliation:
Department of Molecular Biology and Biophysics, University of Connecticut Health Center, Farmington, CT 06030.