Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3'-end processing signals.
Yang, W., Hsu, P.L., Yang, F., Song, J.E., Varani, G.(2018) Nucleic Acids Res 46: 493-503
- PubMed: 29186539
- DOI: https://doi.org/10.1093/nar/gkx1177
- Primary Citation of Related Structures:
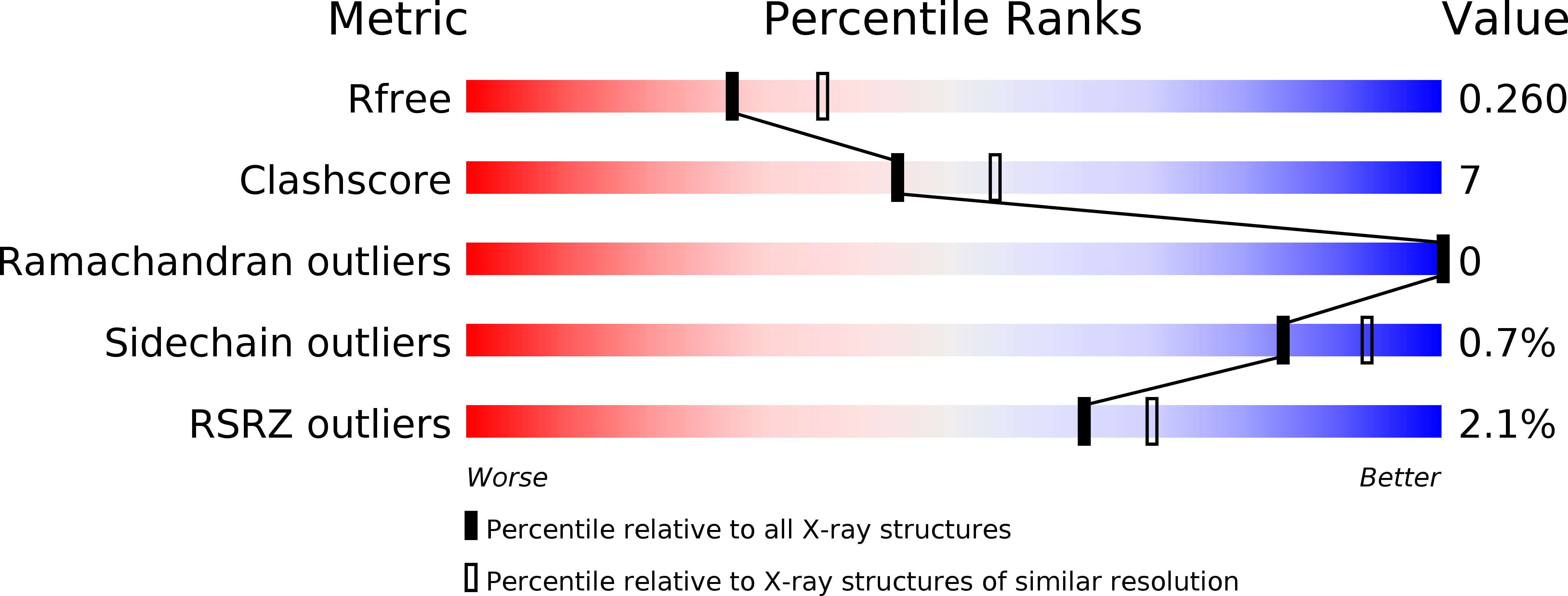
6B3X - PubMed Abstract:
Cleavage stimulation factor (CstF) is a highly conserved protein complex composed of three subunits that recognizes G/U-rich sequences downstream of the polyadenylation signal of eukaryotic mRNAs. While CstF has been identified over 25 years ago, the architecture and contribution of each subunit to RNA recognition have not been fully understood. In this study, we provide a structural basis for the recruitment of CstF-50 to CstF via interaction with CstF-77 and establish that the hexameric assembly of CstF creates a high affinity platform to target various G/U-rich sequences. We further demonstrate that CstF-77 boosts the affinity of the CstF-64 RRM to the RNA targets and CstF-50 fine tunes the ability of the complex to recognize G/U sequences of certain lengths and content.
Organizational Affiliation:
Department of Chemistry, University of Washington, Seattle, WA 98195-1700, USA.