Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
Hirano, T., Fujiwara, T., Niwa, H., Hirano, M., Ohira, K., Okazaki, Y., Sato, S., Umehara, T., Maemoto, Y., Ito, A., Yoshida, M., Kagechika, H.(2018) ChemMedChem 13: 1530-1540
- PubMed: 29882380
- DOI: https://doi.org/10.1002/cmdc.201800233
- Primary Citation of Related Structures:
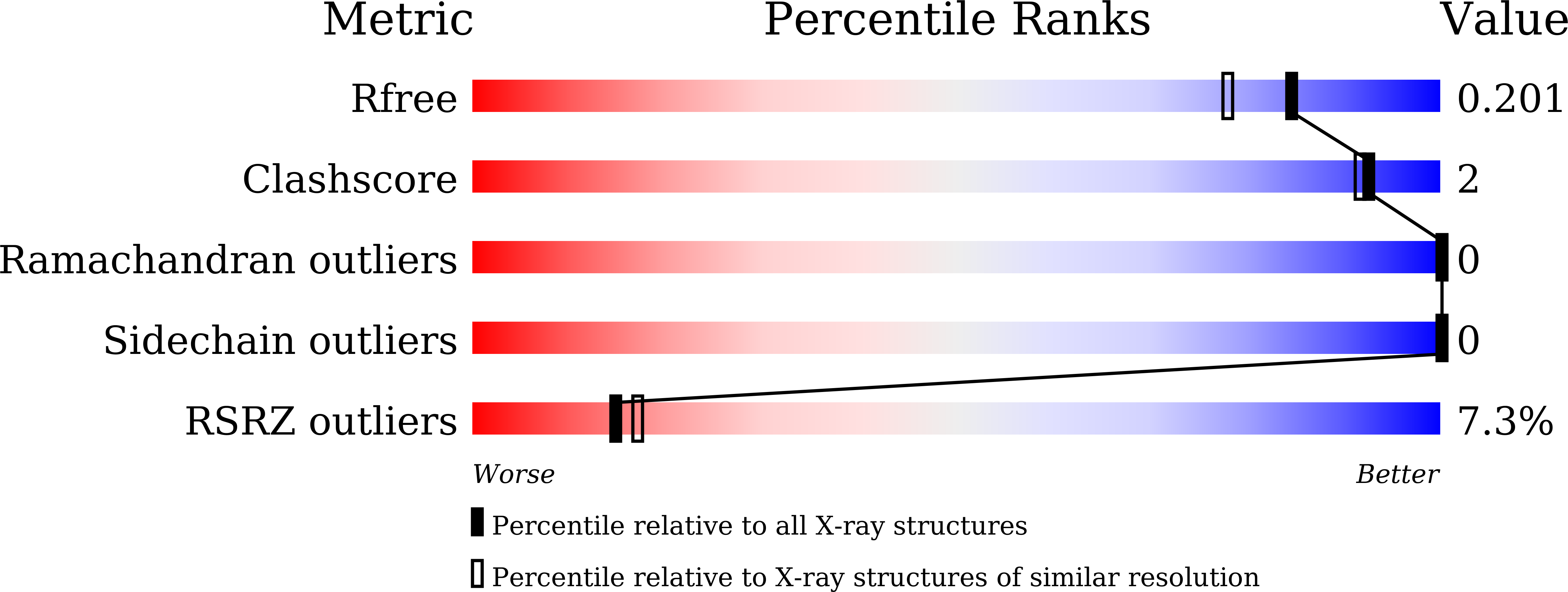
5YLT - PubMed Abstract:
The histone methyltransferase SET7/9 methylates not only histone but also non-histone proteins as substrates, and therefore, SET7/9 inhibitors are considered candidates for the treatment of diseases. Previously, our group identified cyproheptadine, used clinically as a serotonin receptor antagonist and histamine receptor (H1) antagonist, as a novel scaffold of the SET7/9 inhibitor. In this work, we focused on dibenzosuberene as a substructure of cyproheptadine and synthesized derivatives with various functional groups. Among them, the compound bearing a 2-hydroxy group showed the most potent activity. On the other hand, a 3-hydroxy group or another hydrophilic functional group such as acetamide decreased the activity. Structural analysis clarified a rationale for the improved potency only by tightly restricted location and type of the hydrophilic group. In addition, a SET7/9 loop, which was only partially visible in the complex with cyproheptadine, became more clearly visible in the complex with 2-hydroxycyproheptadine. These results are expected to be helpful for further structure-based development of SET7/9 inhibitors.
Organizational Affiliation:
Institute of Biomaterials and Bioengineering, Tokyo Medical and Dental University (TMDU), 2-3-10 Kanda-Surugadai, Chiyoda-ku, Tokyo, 101-0062, Japan.