Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Xiang, Q., Wang, C., Zhang, Y., Xue, X., Song, M., Zhang, C., Li, C., Wu, C., Li, K., Hui, X., Zhou, Y., Smaill, J.B., Patterson, A.V., Wu, D., Ding, K., Xu, Y.(2018) Eur J Med Chem 147: 238-252
- PubMed: 29448139
- DOI: https://doi.org/10.1016/j.ejmech.2018.01.087
- Primary Citation of Related Structures:
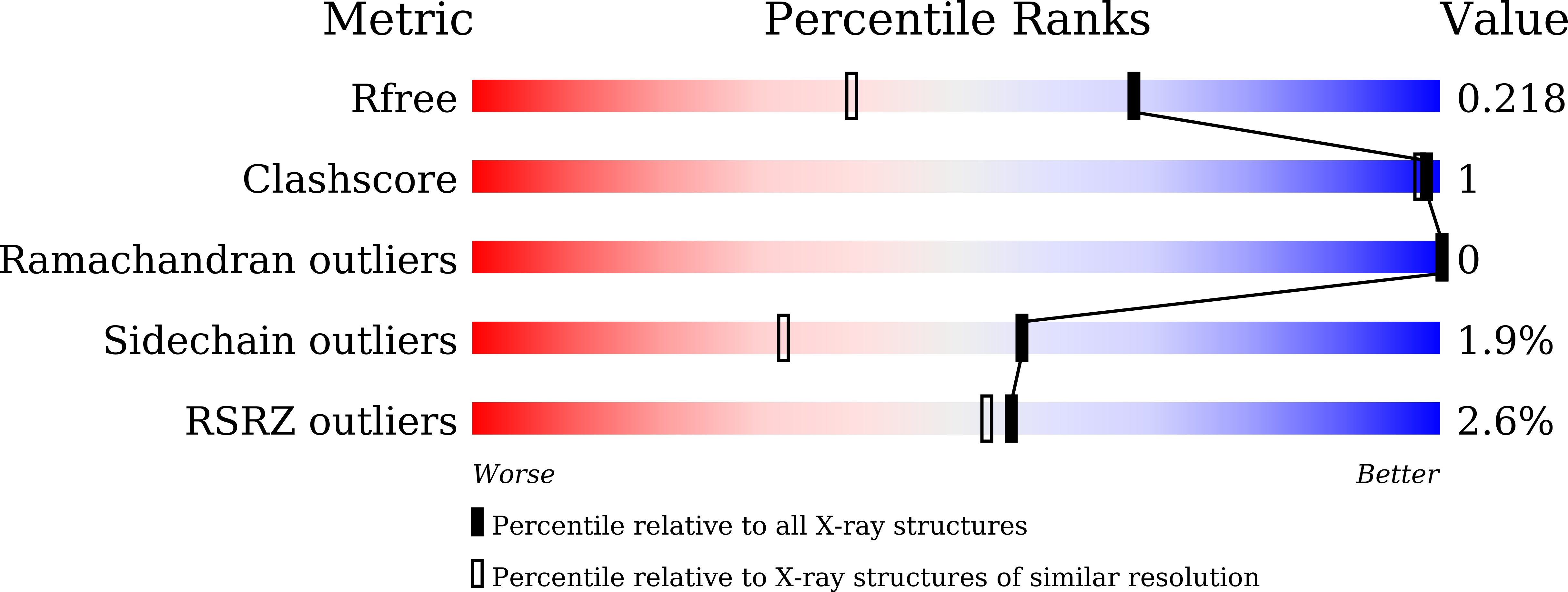
5XXH - PubMed Abstract:
The CREB (cAMP responsive element binding protein) binding protein (CBP) and its homolog EP300 have emerged as new therapeutic targets for the treatment of cancer and inflammatory diseases. Here we report the identification, optimization and evaluation of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 inhibitors starting from fragment-based virtual screening (FBVS). A cocrystal structure of the inhibitor (22e) in complex with CBP provides a solid structural basis for further optimization. The most potent compound 32h binds to the CBP bromodomain and has an IC 50 value of 0.037 μM in the AlphaScreen assay which was 2 times more potent than the reported CBP bromodomain inhibitor SGC-CBP30 in our hands. 32h also exhibit high selectivity for CBP/EP300 over other bromodomain-containing proteins. Notably, the ester derivative (29h) of compound 32h markedly inhibits cell growth in several prostate cancer cell lines including LNCaP, 22Rv1 and LNCaP derived C4-2B. Compound 29h suppresses the mRNA expression of full length AR (AR-FL), AR target genes and other oncogene in LNCaP cells. 29h also reduces the expression of PSA, the biomarker of prostate cancer. CBP/EP300 inhibitor 29h represents a promising lead compound for the development of new therapeutics for the treatment of castration-resistant prostate cancer.
Organizational Affiliation:
Guangdong Provincial Key Laboratory of Biocomputing, Joint School of Life Sciences, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou Medical University, Guangzhou, China; University of Chinese Academy of Sciences, No. 19 Yuquan Road, Beijing 100049, China.