ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Ghai, R., Du, X., Wang, H., Dong, J., Ferguson, C., Brown, A.J., Parton, R.G., Wu, J.W., Yang, H.(2017) Nat Commun 8: 757-757
- PubMed: 28970484
- DOI: https://doi.org/10.1038/s41467-017-00861-5
- Primary Citation of Related Structures:
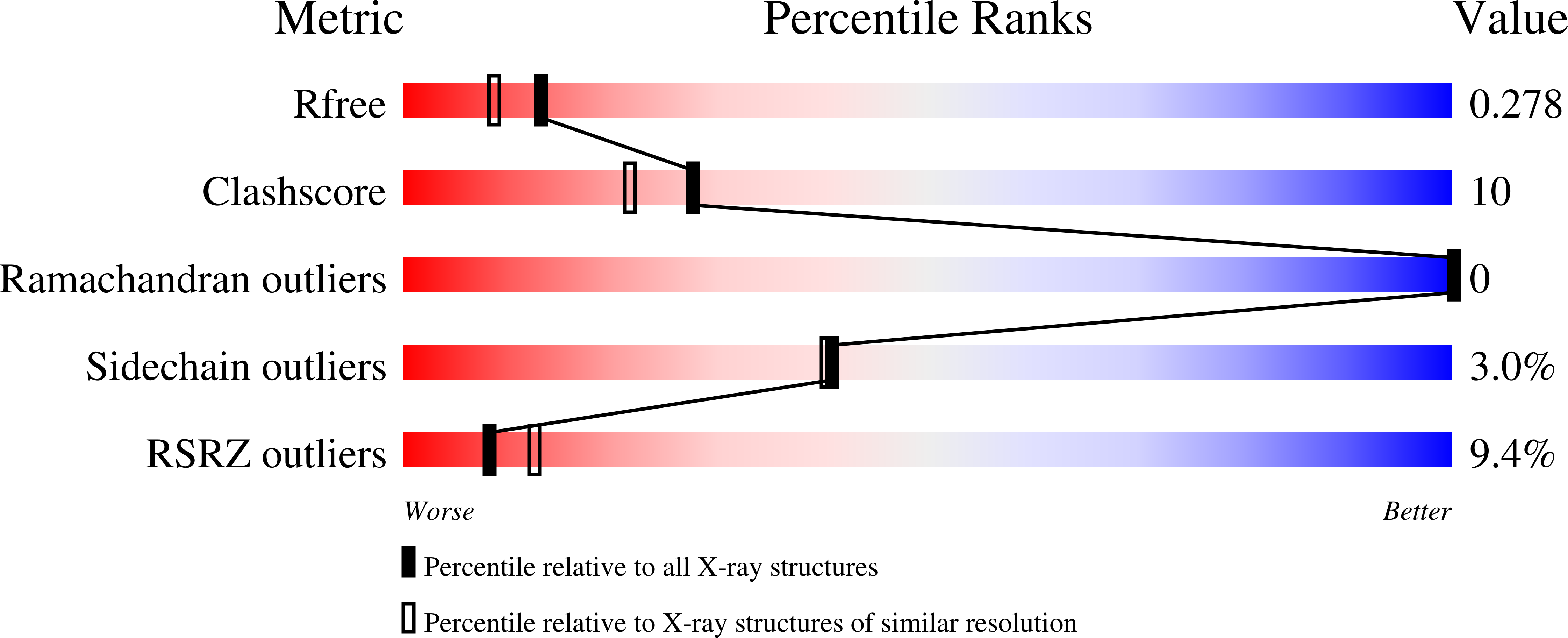
5U77, 5U78 - PubMed Abstract:
ORP5 and ORP8, members of the oxysterol-binding protein (OSBP)-related proteins (ORP) family, are endoplasmic reticulum membrane proteins implicated in lipid trafficking. ORP5 and ORP8 are reported to localize to endoplasmic reticulum-plasma membrane junctions via binding to phosphatidylinositol-4-phosphate (PtdIns(4)P), and act as a PtdIns(4)P/phosphatidylserine counter exchanger between the endoplasmic reticulum and plasma membrane. Here we provide evidence that the pleckstrin homology domain of ORP5/8 via PtdIns(4,5)P 2 , and not PtdIns(4)P binding mediates the recruitment of ORP5/8 to endoplasmic reticulum-plasma membrane contact sites. The OSBP-related domain of ORP8 can extract and transport multiple phosphoinositides in vitro, and knocking down both ORP5 and ORP8 in cells increases the plasma membrane level of PtdIns(4,5)P 2 with little effect on PtdIns(4)P. Overall, our data show, for the first time, that phosphoinositides other than PtdIns(4)P can also serve as co-exchangers for the transport of cargo lipids by ORPs.ORP5/8 are endoplasmic reticulum (ER) membrane proteins implicated in lipid trafficking that localize to ER-plasma membrane (PM) contacts and maintain membrane homeostasis. Here the authors show that PtdIns(4,5)P 2 plays a critical role in the targeting and function of ORP5/8 at the PM.
Organizational Affiliation:
School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney, NSW, 2052, Australia.