PanDDA analysis group deposition
Gray, J.L., Krojer, T., Talon, R., Douangamath, A., Jimenez Antunez, C., Bountra, C., Arrowsmith, C.H., Edwards, A., Brennan, P.E., von Delft, F.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ras-related C3 botulinum toxin substrate 1 | 178 | Homo sapiens | Mutation(s): 0 Gene Names: RAC1, TC25, MIG5 EC: 3.6.5.2 | 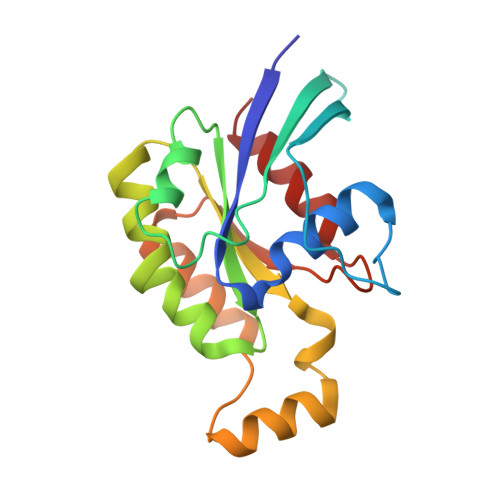 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63000 (Homo sapiens) Explore P63000 Go to UniProtKB: P63000 | |||||
PHAROS: P63000 GTEx: ENSG00000136238 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63000 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Kalirin | 182 | Homo sapiens | Mutation(s): 0 Gene Names: KALRN, DUET, DUO, HAPIP, TRAD EC: 2.7.11.1 | 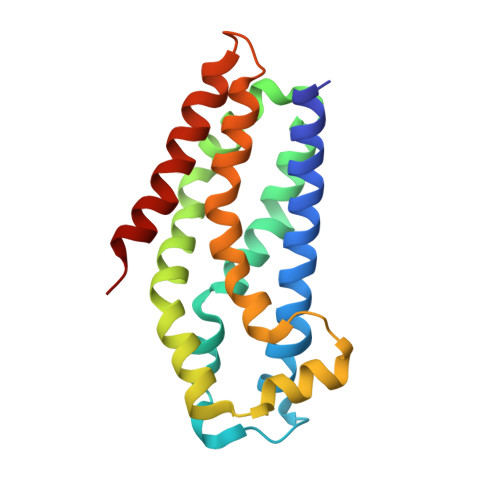 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O60229 (Homo sapiens) Explore O60229 Go to UniProtKB: O60229 | |||||
PHAROS: O60229 GTEx: ENSG00000160145 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O60229 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| N71 Query on N71 | D [auth A] | N-(5-methyl-1,2-oxazol-3-yl)-N'-[2-(phenylsulfonyl)ethyl]urea C13 H15 N3 O4 S XVQVTCYUFZRWGM-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | C [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.308 | α = 90 |
| b = 62.308 | β = 90 |
| c = 340.951 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| REFMAC | phasing |