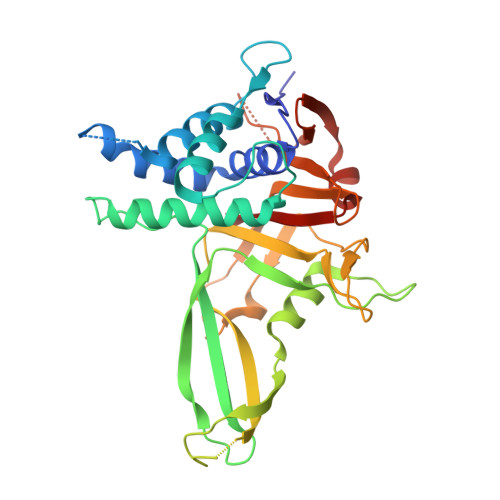
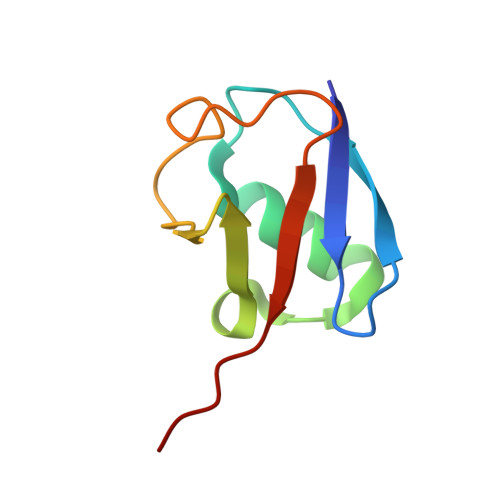
Mechanism and regulation of the Lys6-selective deubiquitinase USP30.
Gersch, M., Gladkova, C., Schubert, A.F., Michel, M.A., Maslen, S., Komander, D.(2017) Nat Struct Mol Biol 24: 920-930
- PubMed: 28945249
- DOI: https://doi.org/10.1038/nsmb.3475
- Primary Citation of Related Structures:
5OHK, 5OHN, 5OHP - PubMed Abstract:
Damaged mitochondria undergo mitophagy, a specialized form of autophagy that is initiated by the protein kinase PINK1 and the ubiquitin E3 ligase Parkin. Ubiquitin-specific protease USP30 antagonizes Parkin-mediated ubiquitination events on mitochondria and is a key negative regulator of mitophagy. Parkin and USP30 both show a preference for assembly or disassembly, respectively, of Lys6-linked polyubiquitin, a chain type that has not been well studied. Here we report crystal structures of human USP30 bound to monoubiquitin and Lys6-linked diubiquitin, which explain how USP30 achieves Lys6-linkage preference through unique ubiquitin binding interfaces. We assess the interplay between USP30, PINK1 and Parkin and show that distally phosphorylated ubiquitin chains impair USP30 activity. Lys6-linkage-specific affimers identify numerous mitochondrial substrates for this modification, and we show that USP30 regulates Lys6-polyubiquitinated TOM20. Our work provides insights into the architecture, activity and regulation of USP30, which will aid drug design against this and related enzymes.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.