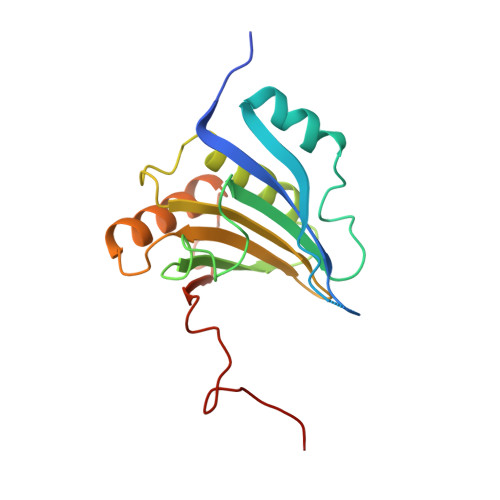
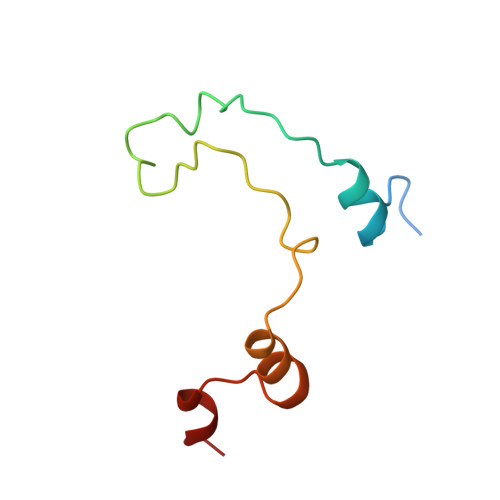
GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Peter, D., Weber, R., Sandmeir, F., Wohlbold, L., Helms, S., Bawankar, P., Valkov, E., Igreja, C., Izaurralde, E.(2017) Genes Dev 31: 1147-1161
- PubMed: 28698298
- DOI: https://doi.org/10.1101/gad.299420.117
- Primary Citation of Related Structures:
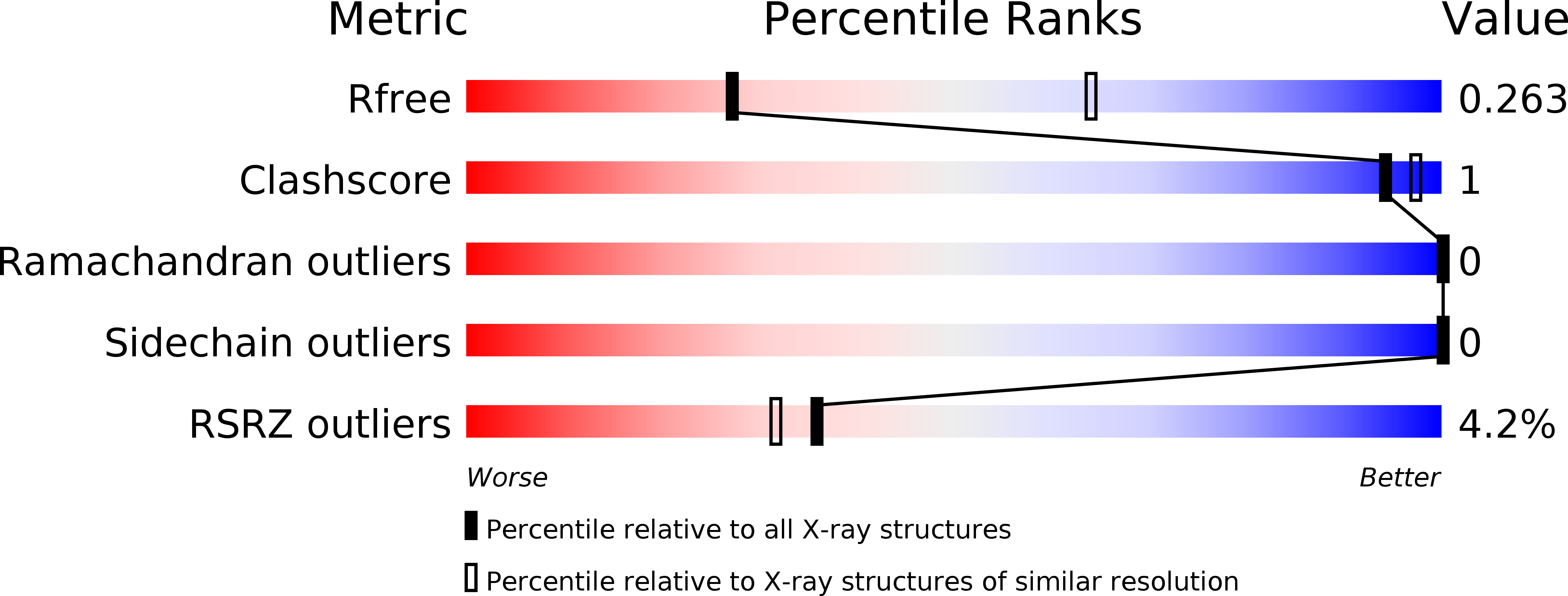
5NVK, 5NVL, 5NVM, 5NVN - PubMed Abstract:
The eIF4E homologous protein (4EHP) is thought to repress translation by competing with eIF4E for binding to the 5' cap structure of specific mRNAs to which it is recruited through interactions with various proteins, including the GRB10-interacting GYF (glycine-tyrosine-phenylalanine domain) proteins 1 and 2 (GIGYF1/2). Despite its similarity to eIF4E, 4EHP does not interact with eIF4G and therefore fails to initiate translation. In contrast to eIF4G, GIGYF1/2 bind selectively to 4EHP but not eIF4E. Here, we present crystal structures of the 4EHP-binding regions of GIGYF1 and GIGYF2 in complex with 4EHP, which reveal the molecular basis for the selectivity of the GIGYF1/2 proteins for 4EHP. Complementation assays in a GIGYF1/2-null cell line using structure-based mutants indicate that 4EHP requires interactions with GIGYF1/2 to down-regulate target mRNA expression. Our studies provide structural insights into the assembly of 4EHP-GIGYF1/2 repressor complexes and reveal that rather than merely facilitating 4EHP recruitment to transcripts, GIGYF1/2 proteins are required for repressive activity.
Organizational Affiliation:
Department of Biochemistry, Max Planck Institute for Developmental Biology, 72076 Tübingen, Germany.