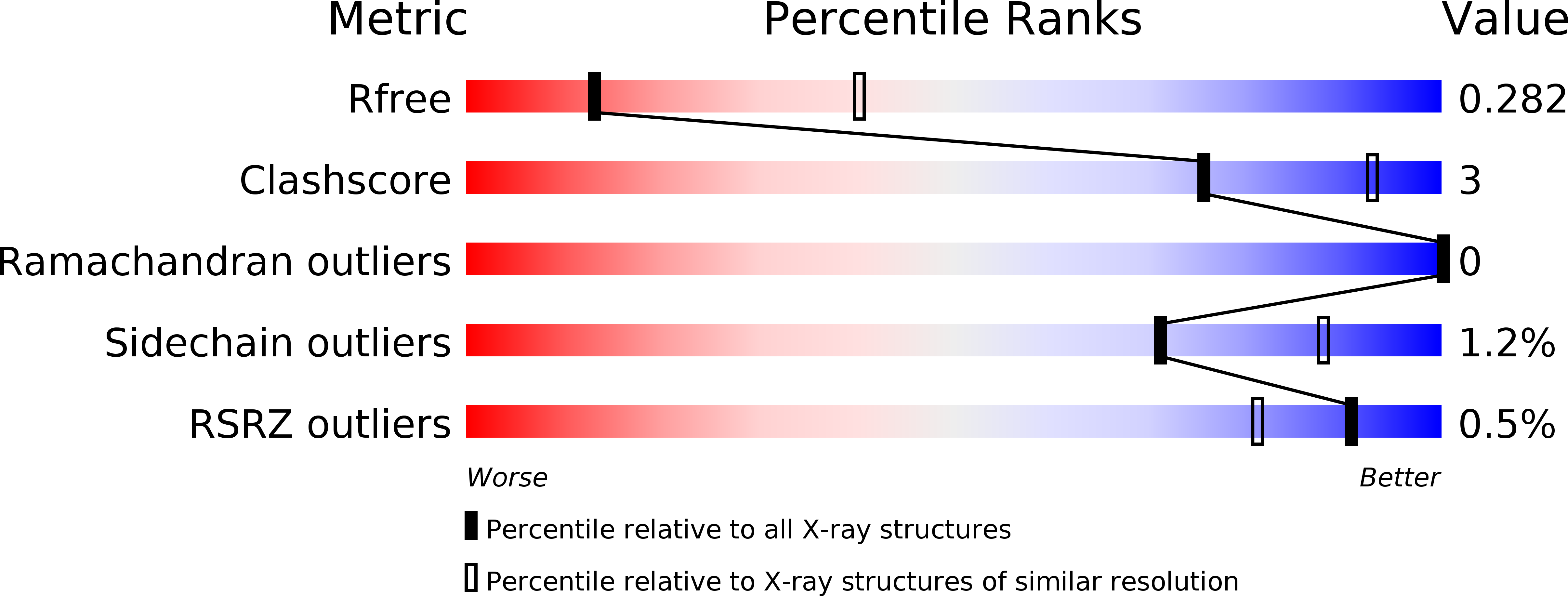
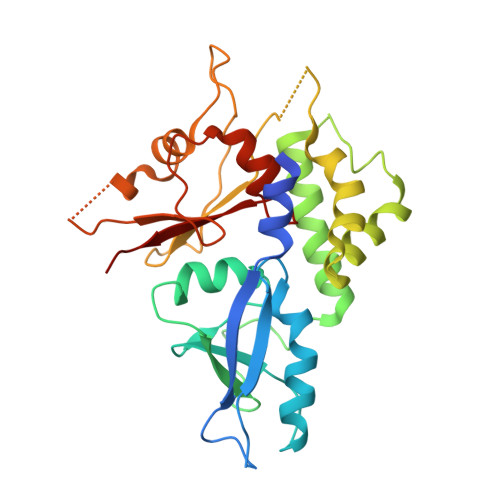
Identification, biochemical characterization and crystallization of the central region of human ATG16L1.
Archna, A., Scrima, A.(2017) Acta Crystallogr F Struct Biol Commun 73: 560-567
- PubMed: 28994404
- DOI: https://doi.org/10.1107/S2053230X17013280
- Primary Citation of Related Structures:
5NPV, 5NPW - PubMed Abstract:
ATG16L1 plays a major role in autophagy. It acts as a molecular scaffold which mediates protein-protein interactions essential for autophagosome formation. The ATG12~ATG5-ATG16L1 complex is one of the key complexes involved in autophagosome formation. Human ATG16L1 comprises 607 amino acids with three functional domains named ATG5BD, CCD and WD40, where the C-terminal WD40 domain represents approximately 50% of the full-length protein. Previously, structures of the C-terminal WD40 domain of human ATG16L1 as well as of human ATG12~ATG5 in complex with the ATG5BD of ATG16L1 have been reported. However, apart from the ATG5BD, no structural information for the N-terminal half, including the CCD, of human ATG16L1 is available. In this study, the authors aimed to structurally characterize the N-terminal half of ATG16L1. ATG16L1 11-307 in complex with ATG5 has been purified and crystallized in two crystal forms. However, both crystal structures revealed degradation of ATG16L1, resulting in crystals comprising only full-length ATG5 and the ATG5BD of ATG16L1. The structures of ATG5-ATG5BD in two novel crystal forms are presented, further supporting the previously observed dimerization of ATG5-ATG16L1. The reported degradation points towards a high instability at the linker region between the ATG5BD and the CCD in ATG16L1. Based on this observation and further biochemical analysis of ATG16L1, a stable 236-amino-acid subfragment comprising residues 72-307 of the N-terminal half of ATG16L1, covering the residual, so far structurally uncharacterized region of human ATG16L1, was identified. Here, the identification, purification, biochemical characterization and crystallization of the proteolytically stable ATG16L1 72-307 subfragment are reported.
Organizational Affiliation:
Structural Biology of Autophagy Group, Department of Structure and Function of Proteins, Helmholtz Centre for Infection Research, Inhoffenstrasse 7, 38124 Braunschweig, Germany.