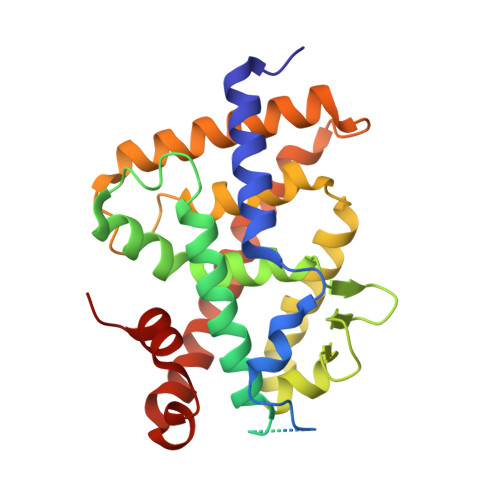
Structural analysis and biological activities of BXL0124, a gemini analog of vitamin D.
Belorusova, A.Y., Suh, N., Lee, H.J., So, J.Y., Maehr, H., Rochel, N.(2017) J Steroid Biochem Mol Biol 173: 69-74
- PubMed: 27650654
- DOI: https://doi.org/10.1016/j.jsbmb.2016.09.015
- Primary Citation of Related Structures:
5LGA - PubMed Abstract:
Gemini analogs of calcitriol, characterized by the extension of the C21-methyl group of calcitriol with a second chain, act as agonists of the vitamin D receptor (VDR). This second side chain of gemini is accommodated in a new cavity inside the VDR created by the structural rearrangement of the protein core. The resulting conformational change preserves the active state of the receptor and bestows gemini compounds with biological activities that exceed those of calcitriol. Of particular interest are gemini's anti-cancer properties, and in this study we demonstrate anti-proliferative and tumor-reducing abilities of BXL0124 and BXL0097, differing only by the presence or absence, respectively, of the methylene group on the A ring. BXL0124 acts as a more potent VDR agonist than its 19-nor counterpart by activating VDR-mediated transcription at lower concentrations. In a similar manner, BXL0124 is more active than BXL0097 in growth inhibition of breast cancer cells and reduction of tumor volume. Structural comparisons of BXL0097 and BXL0124, as their VDR complexes, explain the elevated activity of the latter.
Organizational Affiliation:
Department of Integrated Structural Biology, IGBMC (Institute of Genetics and of Molecular and Cellular Biology), 1 rue Laurent Fries, Illkirch, France; Centre National de la Recherche Scientifique (CNRS) UMR 7104, Illkirch, France; Institut National de la Santé et de la Recherche Médicale (INSERM) U964, Illkirch, France; Université de Strasbourg, Strasbourg, France.