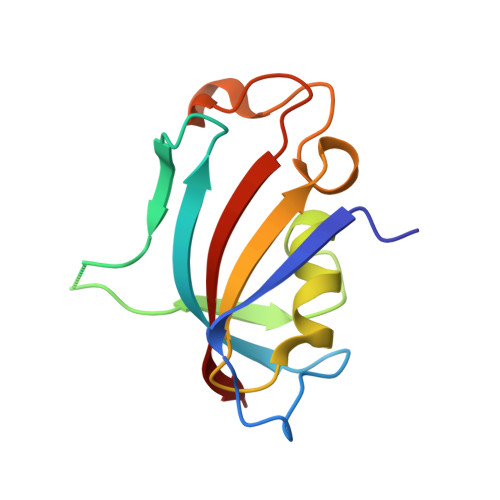
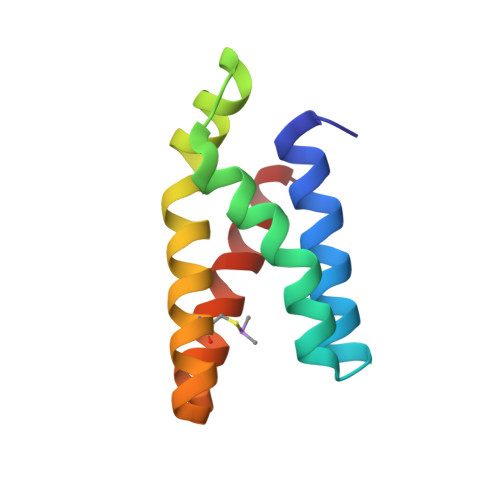
Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Lee, S.Y., Lee, H., Lee, H.K., Lee, S.W., Ha, S.C., Kwon, T., Seo, J.K., Lee, C., Rhee, H.W.(2016) ACS Cent Sci 2: 506-516
- PubMed: 27610411
- DOI: https://doi.org/10.1021/acscentsci.6b00137
- Primary Citation of Related Structures:
5GPG - PubMed Abstract:
Mammalian target of rapamycin (mTOR) signaling is a core pathway in cellular metabolism, and control of the mTOR pathway by rapamycin shows potential for the treatment of metabolic diseases. In this study, we employed a new proximity biotin-labeling method using promiscuous biotin ligase (pBirA) to identify unknown elements in the rapamycin-induced interactome on the FK506-rapamycin binding (FRB) domain in living cells. FKBP25 showed the strongest biotin labeling by FRB-pBirA in the presence of rapamycin. Immunoprecipitation and immunofluorescence experiments confirmed that endogenous FKBP25 has a rapamycin-induced physical interaction with the FRB domain. Furthermore, the crystal structure of the ternary complex of FRB-rapamycin-FKBP25 was determined at 1.67-Å resolution. In this crystal structure we found that the conformational changes of FRB generate a hole where there is a methionine-rich space, and covalent metalloid coordination was observed at C2085 of FRB located at the bottom of the hole. Our results imply that FKBP25 might have a unique physiological role related to metallomics in mTOR signaling.
Organizational Affiliation:
Department of Chemistry, Department of Biological Sciences, UNIST Central Research Facilities (UCRF), and Deparment of Biomedical Engineering, Ulsan National Institute of Science and Technology (UNIST) , Ulsan 44919, Korea.