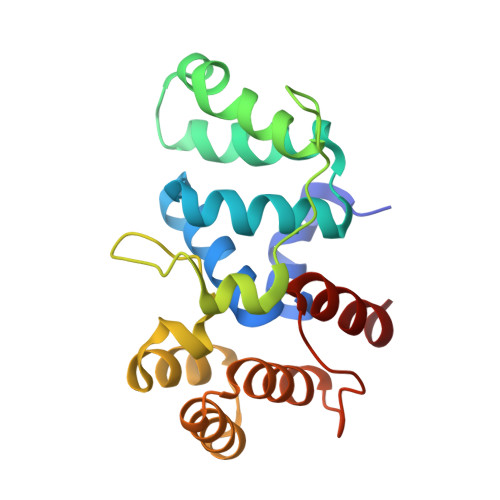
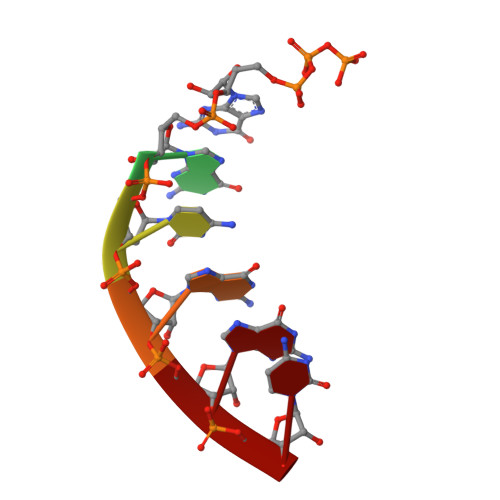
Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome.
Baranovskiy, A.G., Babayeva, N.D., Zhang, Y., Gu, J., Suwa, Y., Pavlov, Y.I., Tahirov, T.H.(2016) J Biol Chem 291: 10006-10020
- PubMed: 26975377
- DOI: https://doi.org/10.1074/jbc.M116.717405
- Primary Citation of Related Structures:
5EXR, 5F0Q, 5F0S - PubMed Abstract:
The human primosome, a 340-kilodalton complex of primase and DNA polymerase α (Polα), synthesizes chimeric RNA-DNA primers to be extended by replicative DNA polymerases δ and ϵ. The intricate mechanism of concerted primer synthesis by two catalytic centers was an enigma for over three decades. Here we report the crystal structures of two key complexes, the human primosome and the C-terminal domain of the primase large subunit (p58C) with bound DNA/RNA duplex. These structures, along with analysis of primase/polymerase activities, provide a plausible mechanism for all transactions of the primosome including initiation, elongation, accurate counting of RNA primer length, primer transfer to Polα, and concerted autoregulation of alternate activation/inhibition of the catalytic centers. Our findings reveal a central role of p58C in the coordinated actions of two catalytic domains in the primosome and ultimately could impact the design of anticancer drugs.
Organizational Affiliation:
From the Eppley Institute for Research in Cancer and Allied Diseases, Fred & Pamela Buffett Cancer Center and.