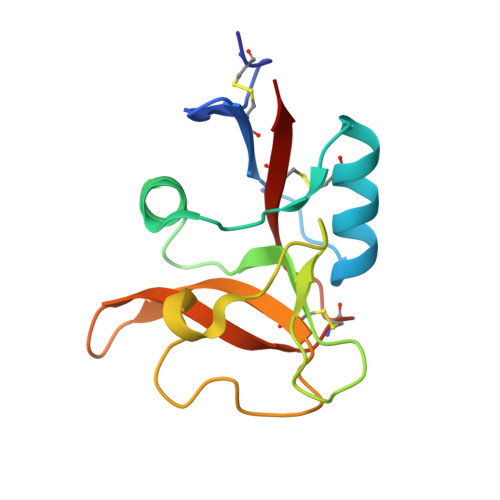
Crystal structure of human dendritic cell inhibitory receptor C-type lectin domain reveals the binding mode with N-glycan
Nagae, M., Ikeda, A., Hanashima, S., Kojima, T., Matsumoto, N., Yamamoto, K., Yamaguchi, Y.(2016) FEBS Lett 590: 1280-1288
- PubMed: 27015765
- DOI: https://doi.org/10.1002/1873-3468.12162
- Primary Citation of Related Structures:
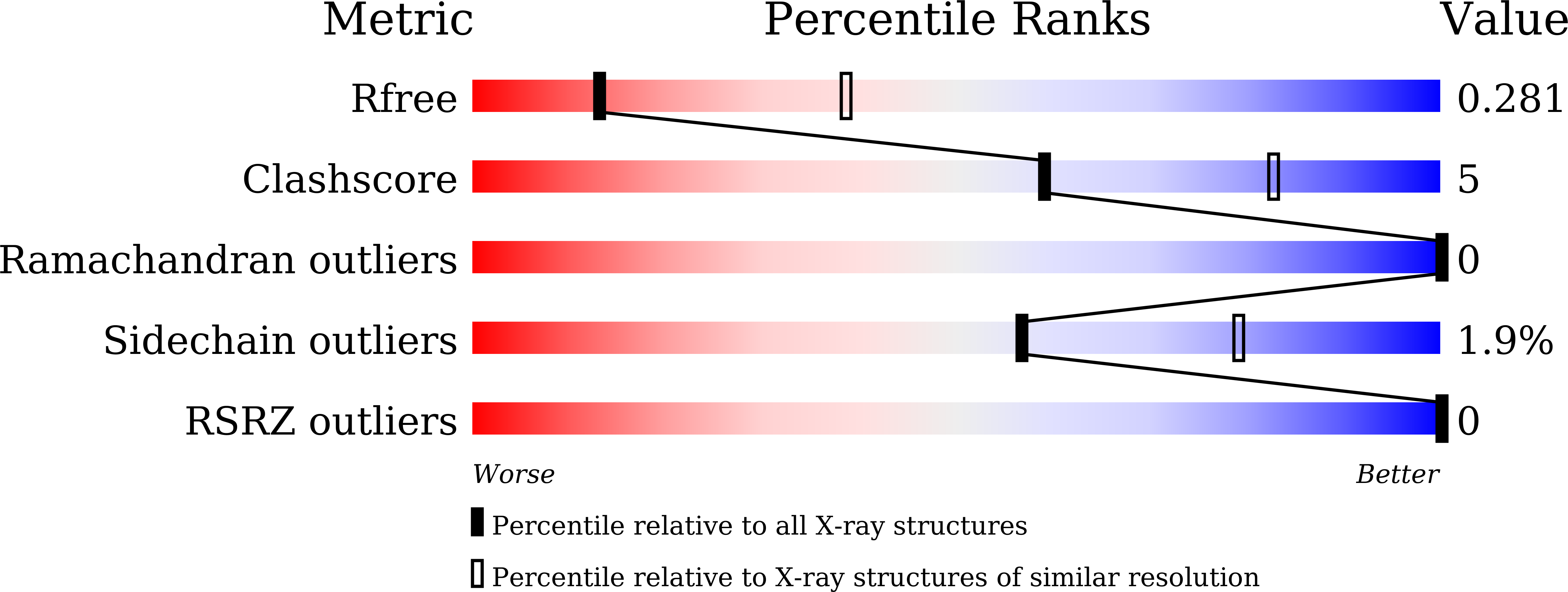
5B1W, 5B1X - PubMed Abstract:
Human dendritic cell inhibitory receptor (DCIR) is a C-type lectin receptor expressed in classical dendritic cells and accepts several oligosaccharide ligands including N-glycans. Here, we report the crystal structures of human DCIR C-type lectin domains in the absence and presence of a branched N-glycan unit. The domain has a typical C-type lectin fold and two bound calcium ions. In the ligand-bound form, the disaccharide unit (GlcNAcβ1-2Man) acceptably fits the electron density map, indicating that it forms the main epitope. The recognition of the nonterminal N-glycan unit explains the relatively broad specificity of this lectin.
Organizational Affiliation:
Structural Glycobiology Team, Systems Glycobiology Research Group, RIKEN-Max Planck Joint Research Center, RIKEN Global Research Cluster, Wako, Saitama, Japan.