Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
Kung, P.P., Bingham, P., Brooun, A., Collins, M., Deng, Y.L., Dinh, D., Fan, C., Gajiwala, K.S., Grantner, R., Gukasyan, H.J., Hu, W., Huang, B., Kania, R., Kephart, S.E., Krivacic, C., Kumpf, R.A., Khamphavong, P., Kraus, M., Liu, W., Maegley, K.A., Nguyen, L., Ren, S., Richter, D., Rollins, R.A., Sach, N., Sharma, S., Sherrill, J., Spangler, J., Stewart, A.E., Sutton, S., Uryu, S., Verhelle, D., Wang, H., Wang, S., Wythes, M., Xin, S., Yamazaki, S., Zhu, H., Zhu, J., Zehnder, L., Edwards, M.(2018) J Med Chem 61: 650-665
- PubMed: 29211475
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01375
- Primary Citation of Related Structures:
4W2R, 6B3W - PubMed Abstract:
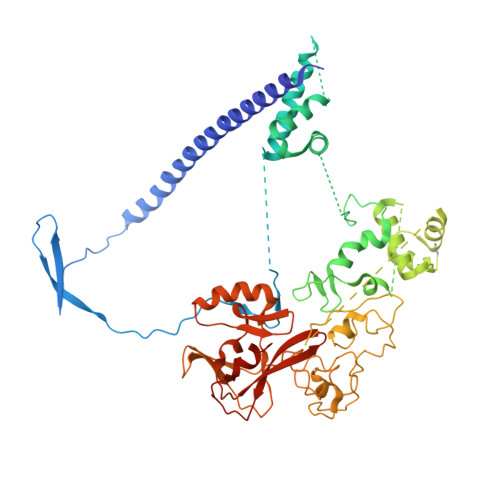
A new series of lactam-derived EZH2 inhibitors was designed via ligand-based and physicochemical-property-based strategies to address metabolic stability and thermodynamic solubility issues associated with previous lead compound 1. The new inhibitors incorporated an sp 3 hybridized carbon atom at the 7-position of the lactam moiety present in lead compound 1 as a replacement for a dimethylisoxazole group. This transformation enabled optimization of the physicochemical properties and potency compared to compound 1. Analysis of relationships between calculated log D (clogD) values and in vitro metabolic stability and permeability parameters identified a clogD range that afforded an increased probability of achieving favorable ADME data in a single molecule. Compound 23a exhibited the best overlap of potency and pharmaceutical properties as well as robust tumor growth inhibition in vivo and was therefore advanced as a development candidate (PF-06821497). A crystal structure of 23a in complex with the three-protein PRC2 complex enabled understanding of the key structural features required for optimal binding.
Organizational Affiliation:
WuXi AppTec , 288 Fute Zhong Road, Waigaoqiao Free Trade Zone, Shanghai 200131, China.