DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Xu, C., Liu, K., Lei, M., Yang, A., Li, Y., Hughes, T.R., Min, J.(2018) Structure 26: 85-95.e3
- PubMed: 29276034
- DOI: https://doi.org/10.1016/j.str.2017.11.022
- Primary Citation of Related Structures:
4NW3, 4O64, 4PZI, 4Z3C, 5VC9, 5W9Q, 5W9S, 6ASB, 6ASD - PubMed Abstract:
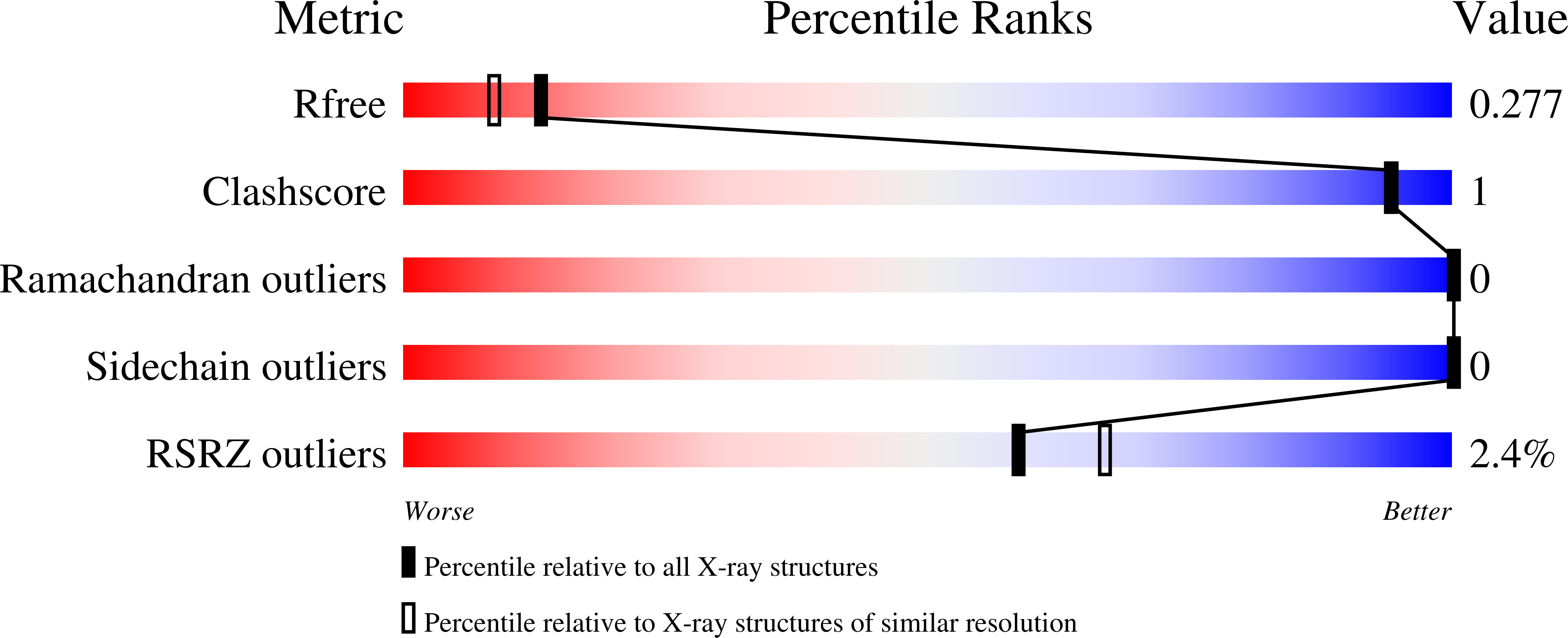
The CXXC domain, first identified as the reader of unmodified CpG dinucleotide, plays important roles in epigenetic regulation by targeting various activities to CpG islands. Here we systematically measured and compared the DNA-binding selectivities of all known human CXXC domains by different binding assays, and complemented the existing function-based classification of human CXXC domains with a classification based on their DNA selectivities. Through a series of crystal structures of CXXC domains with DNA ligands, we unravel the molecular mechanisms of how these CXXC domains, including single CXXC domains and tandem CXXC-PHD domains, recognize distinct DNA ligands, which further supports our classification of human CXXC domains and also provides insights into selective recruitment of chromatin modifiers to their respective targets via CXXC domains recognizing different genomic DNA sequences. Our study facilitates the understanding of the relationship between the DNA-binding specificities of the CXXC proteins and their biological functions.
Organizational Affiliation:
Structural Genomics Consortium, University of Toronto, Toronto, ON M5G 1L7, Canada.