Molecular Determinants of Agonist and Antagonist Signaling through the IL-36 Receptor.
Gunther, S., Sundberg, E.J.(2014) J Immunol 193: 921-930
- PubMed: 24935927
- DOI: https://doi.org/10.4049/jimmunol.1400538
- Primary Citation of Related Structures:
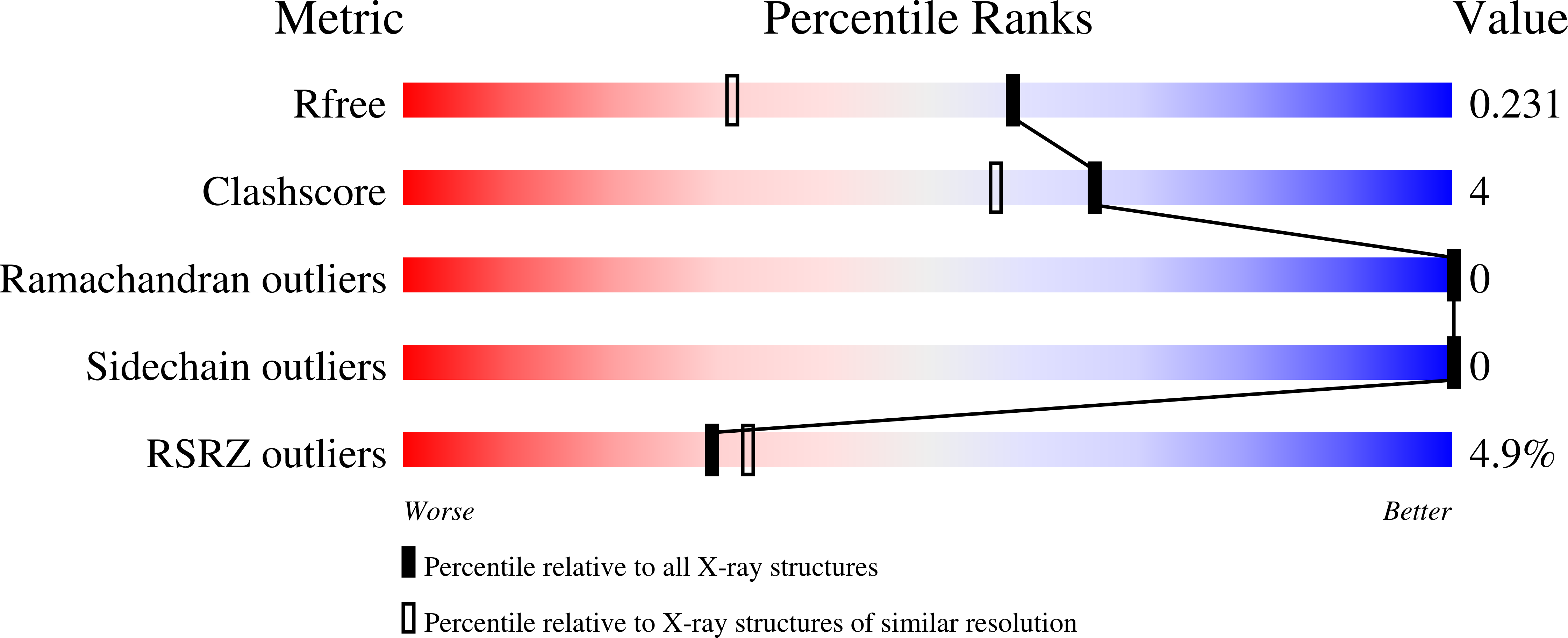
4IZE, 4P0J, 4P0K, 4P0L - PubMed Abstract:
The IL-1 family consists of 11 cytokines that control a complex network of proinflammatory signals critical for regulating immune responses to infections. They also play a central role in numerous chronic inflammatory disorders. Accordingly, inhibiting the activities of these cytokines is an important therapeutic strategy for treating autoimmune diseases and lymphomas. Agonist cytokines in the IL-1 family activate signaling by binding their cognate receptor and then recruiting a receptor accessory protein. Conversely, antagonist cytokines bind their cognate receptor but prohibit recruitment of receptor accessory protein, which precludes functional signaling complexes. The IL-36 subfamily of cytokines is the most diverse, including three agonists and at least one antagonist, and is the least well-characterized group within this family. Signaling through the IL-36 receptor directly stimulates dendritic cells and primes naive CD4 T cells for Th1 responses. Appropriately balanced IL-36 signaling is a critical determinant of skin and lung health. IL-36 signaling has been presumed to function analogously to IL-1 signaling. In this study, we have defined molecular determinants of agonist and antagonist signaling through the IL-36 receptor. We present the crystal structure of IL-36γ, which, to our knowledge, is the first reported structure of an IL-36 agonist. Using this structure as a guide, we designed a comprehensive series of IL-36 agonist/antagonist chimeric proteins for which we measured binding to the IL-36 receptor/IL-1 receptor accessory protein complex and functional activation and inhibition of signaling. Our data reveal how the fine specificity of IL-36 signaling is distinct from that of IL-1.
Organizational Affiliation:
Institute of Human Virology, University of Maryland School of Medicine, Baltimore, MD 21201;