Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 4. Exploration of a novel binding pocket.
Hong, F.T., Norman, M.H., Ashton, K.S., Bartberger, M.D., Chen, J., Chmait, S., Cupples, R., Fotsch, C., Jordan, S.R., Lloyd, D.J., Sivits, G., Tadesse, S., Hale, C., St Jean, D.J.(2014) J Med Chem 57: 5949-5964
- PubMed: 25001129
- DOI: https://doi.org/10.1021/jm5001979
- Primary Citation of Related Structures:
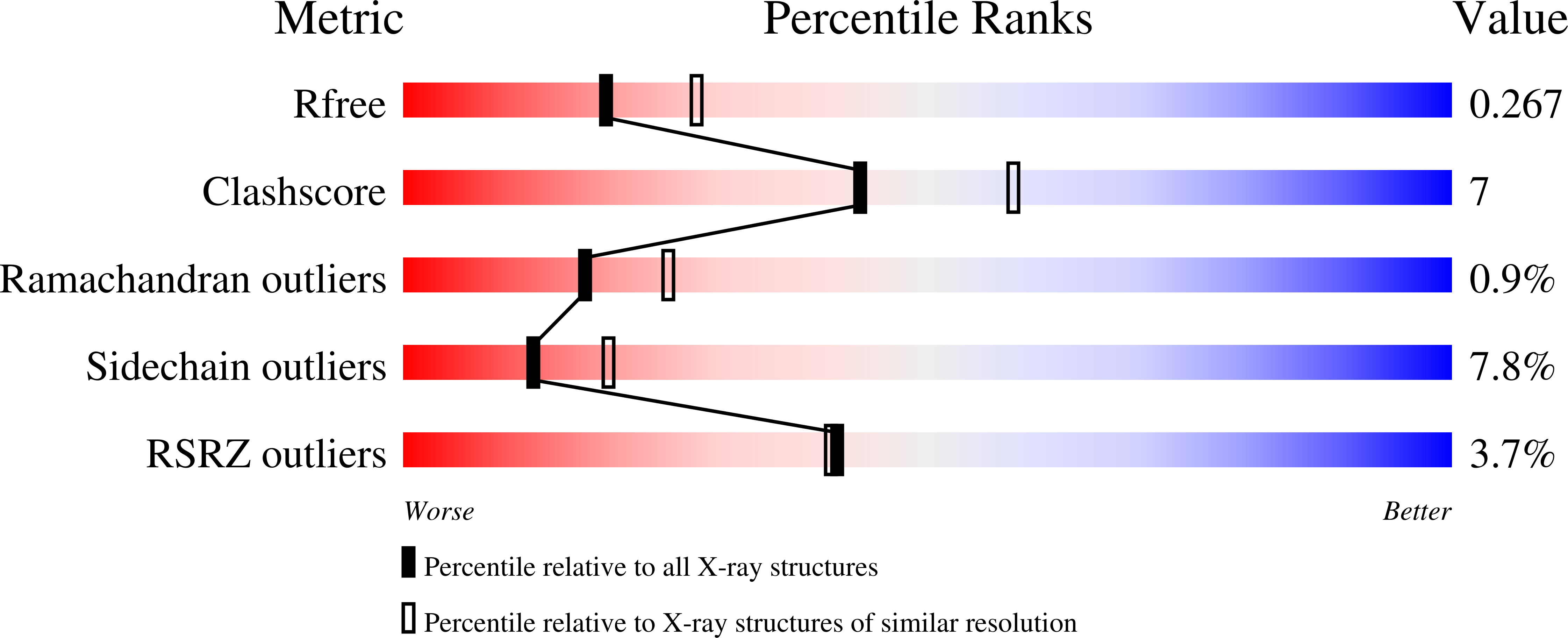
4OLH - PubMed Abstract:
Structure-activity relationship investigations conducted at the 5-position of the N-pyridine ring of a series of N-arylsulfonyl-N'-2-pyridinyl-piperazines led to the identification of a novel bis-pyridinyl piperazine sulfonamide (51) that was a potent disruptor of the glucokinase-glucokinase regulatory protein (GK-GKRP) interaction. Analysis of the X-ray cocrystal of compound 51 bound to hGKRP revealed that the 3-pyridine ring moiety occupied a previously unexplored binding pocket within the protein. Key features of this new binding mode included forming favorable contacts with the top face of the Ala27-Val28-Pro29 ("shelf region") as well as an edge-to-face interaction with the Tyr24 side chain. Compound 51 was potent in both biochemical and cellular assays (IC50=0.005 μM and EC50=0.205 μM, respectively) and exhibited acceptable pharmacokinetic properties for in vivo evaluation. When administered to db/db mice (100 mg/kg, po), compound 51 demonstrated a robust pharmacodynamic effect and significantly reduced blood glucose levels up to 6 h postdose.
Organizational Affiliation:
Departments of Therapeutic Discovery-Medicinal Chemistry, ‡ Therapeutic Discovery-Molecular Structure, §Pharmacokinetics and Drug Metabolism, and ∥Metabolic Disorders, Amgen, Inc. , One Amgen Center Drive, Thousand Oaks, California 91320-1799, United States.